[Paper] Translation of a Paper on COVID-19 and Genetics #1
![[Paper] Translation of a Paper on COVID-19 and Genetics #1](https://humedit.com/wp-content/uploads/paper.jpg)
Table of Contents
Genetic Mechanisms of Severe COVID-19
Received: September 27, 2020
Accepted: November 30, 2020
Accelerated article preview published
Online publication date: December 11, 2020
To cite this article: Pairo-Castineira, E. et al.
Genetic mechanisms of severe COVID-19.
Nature https://doi.org/10.1038/
s41586-020-03065-y (2020).
Erola Pairo-Castineira1,2,325, Sara Clohisey1,325, Lucija Klaric2,325, Andrew D. Bretherick2,325, Konrad Rawlik1,325, Dorota Pasko3, Susan Walker3, Nick Parkinson1, Max Head Fourman1, Clark D. Russell1,4, James Furniss1, Anne Richmond2, Elvina Gountouna5, Nicola Wrobel6, David Harrison7, Bo Wang1, Yang Wu8, Alison Meynert2, Fiona Griffiths1, Wilna Oosthuyzen1, Athanasios Kousathanas3, Loukas Moutsianas3, Zhijian Yang9, Ranran Zhai9, Chenqing Zheng9, Graeme Grimes2, Rupert Beale10, Jonathan Millar1, Barbara Shih1, Sean Keating11, Marie Zechner1, Chris Haley1, David J. Porteous5, Caroline Hayward2,5, Jian Yang12,13, Julian Knight14, Charlotte Summers15, Manu Shankar-Hari16,17, Paul Klenerman14, Lance Turtle18, Antonia Ho19, Shona C. Moore18, Charles Hinds20, Peter Horby21, Alistair Nichol22,23,24, David Maslove25, Lowell Ling26, Danny McAuley27,28, Hugh Montgomery29, Timothy Walsh11, Alex Pereira30, Alessandra Renieri31,32, The GenOMICC Investigators*, The ISARIC4C Investigators*, The COVID-19 Human Genetics Initiative*, 23andMe Investigators*, BRACOVID Investigators*, Gen-COVID Investigators*, Xia Shen9,33,34, Chris P. Ponting2, Angie Fawkesv6, Albert Tenesa1,2,33, Mark Caulfield3,20, Richard Scott3,35, Kathy Rowan7, Lee Murphy6, Peter J. M. Openshaw36,37, Malcolm G. Semple18,38, Andrew Law1, Veronique Vitart2, James F. Wilson2,33 & J. Kenneth Baillie1,2,11 ✉
Severe illness caused by Covid-19 involves host-mediated lung inflammation, which increases mortality rates. Host genetic variations associated with severe disease may identify targets for therapeutic development. Here, we report the results of a GenOMICC (Genetics Of Mortality In Critical Care) genome-wide association study (GWAS) in 2244 critically ill Covid-19 patients from 208 UK intensive care units (ICUs). We identify and replicate novel genome-wide significant associations at chr12q24.13 (rs10735079, p = 1.65×10-8) within a gene cluster encoding antiviral restriction enzyme activators (OAS1, OAS2, OAS3), chr19p13.2 (rs2109069, p = 2.3×10-12) near TYK2, chr19p13.3 (rs2109069, p = 3.98×10-12) within DPP9, and chr21q22.1 (rs2236757, p = 4.99×10-8) within the interferon receptor gene IFNAR2. These associations identify potential targets for repurposing approved drugs. Using Mendelian randomization, we found evidence supporting a causal relationship between low expression of IFNAR2 and high expression of TYK2 with life-threatening disease. Transcriptome-wide association in lung tissue revealed that high expression of the monocyte/macrophage chemotactic receptor CCR2 is associated with severe Covid-19. Our results identify strong genetic signals related to key host antiviral defense mechanisms and mediators of inflammatory organ damage in Covid-19. Both mechanisms may be amenable to targeted therapy with existing drugs. Large-scale randomized clinical trials are essential before any changes in clinical practice.
Severe illness from Covid-19 is partly caused by inflammatory damage affecting the lungs and pulmonary vasculature. There are at least two distinct biological components to the risk of death: susceptibility to viral infection and propensity to develop harmful lung inflammation. Susceptibility to life-threatening infections and immune-mediated diseases is highly heritable. Specifically, susceptibility to respiratory viruses such as influenza is known to be heritable and associated with specific genetic variants. In Covid-19, a single locus at 3p21.31 has been repeatedly associated with hospitalization. As with other viral diseases, loss-of-function variants affecting essential immune processes, such as TLR7 and associated genes, have been identified in young people with severe disease. Genome-wide studies have the potential to uncover entirely novel molecular mechanisms of severe disease in Covid-19, which may provide therapeutic targets to modulate the host immune response and improve survival.
There is now strong evidence that severe illness caused by Covid-19 is qualitatively different from mild or moderate disease, even among hospitalized patients. Patterns of symptoms vary, and there are distinct disease phenotypes with markedly different responses to immunosuppressive therapy. In patients without respiratory failure, treatment may cause harm, whereas in patients with severe respiratory failure, corticosteroid use yields substantial benefits. Based on this, we consider that patients with severe Covid-19 respiratory failure have a distinct pathophysiology. In the UK, patients admitted to intensive care for life-threatening illness are relatively homogeneous, typically presenting with severe hypoxic respiratory failure. These patients' active disease processes respond markedly to corticosteroid therapy and are characterized by lung inflammation, including diffuse alveolar damage, pulmonary macrophage/monocyte influx, mononuclear cell pulmonary vasculitis, and microthrombosis. Host-directed therapies have long been desired for treating severe disease caused by respiratory viruses. Identification of loci associated with susceptibility to Covid-19 may lead to specific targets for drug repurposing or development.
The GenOMICC (Genetics Of Mortality In Critical Care, genomicc.org) study has been recruiting patients with severe syndromes such as influenza, sepsis, and emerging infections for five years. To better understand host mechanisms leading to life-threatening Covid-19, we conducted a genome-wide association study comparing critically ill Covid-19 patients with controls from a UK population genetics study.
Results
Severe cases were recruited through the GenOMICC study in 208 UK intensive care units, and hospitalized cases were recruited through the ISARIC Coronavirus Clinical Characterization Consortium (4C) study. DNA was extracted from high-quality whole blood and array-based genome-wide genotyping obtained from 2734 unique individuals (Materials and Methods). Genetic ancestry was inferred using principal component analysis and individuals from the 1000 Genomes Project as the population reference (Materials and Methods). After quality control and matching to ancestry groups, 2244 individuals were included in the GWAS analysis. The clinical and demographic characteristics of these cases are shown in Extended Data Table 1. Additional clinical details for a subset of 1069 cases with available supplementary data are shown in Supplementary Figures 7-13 and Supplementary Table 2. The cases were critically representative of the UK population with severe illness. In this multi-ancestry cohort, imputation was performed using the TOPMed reference panel.
Ancestry-matched controls were selected from the large population-based cohort UK Biobank (five controls for each case). Controls known to have tested positive for Covid-19 were excluded. The inevitable presence of individuals in the control group who might exhibit severe illness phenotypes if exposed to SARS-CoV-2 is expected to bias associations towards the null. GWAS was performed separately for each ancestry group using logistic regression in PLINK, adjusting for age, sex, decile of postal code deprivation, and ancestry principal components. In addition to several standard filters to minimize spurious associations (Materials and Methods), a subset of 1613 cases underwent whole-genome sequencing to filter out poorly called or imputed variants. Therefore, of the 4,469,187 imputed variants that passed other quality control filters after GWAS, 83,937 were filtered out. There was a high level of residual inflation in the South Asian and East Asian ancestry groups, making results from these subgroups unreliable (Extended Data Fig. 1, Supplementary Fig. 4). The largest ancestry group included 1676 individuals of European ancestry (EUR), which was used for the primary analysis presented below.
GWAS Results
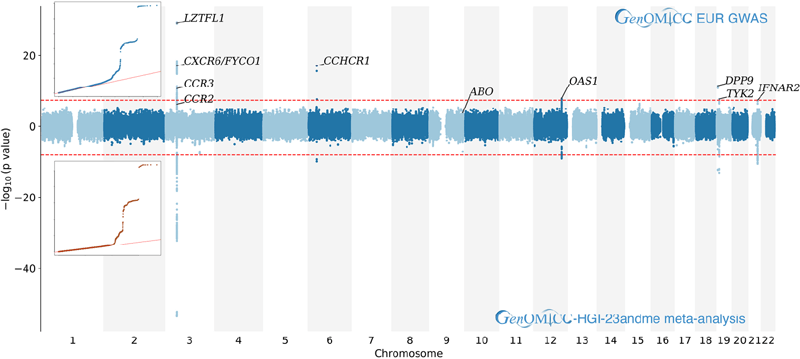
In the primary analysis (GenOMICC European cases vs. UK Biobank controls), 15 independent association signals were genome-wide significant at p<5×10-8 after linkage disequilibrium-based clumping (Supplementary Fig. 1). Of these, eight were successfully validated in GWAS using two independent population genetic studies (100,000 Genomes and Generation Scotland) as controls (Table 1), leading to their replication. The sex-specific GWAS in this group did not reveal any sex-specific associations (Supplementary Table 1). No additional associations were identified in the trans-ethnic meta-analysis (Supplementary Fig. 3).
Replication
Due to the lack of sufficiently large studies on severe Covid-19 disease, reproducibility was sought through a meta-analysis of data from the Covid-19 Host Genetics Initiative (HGI, mixed ancestry) involving 2415 hospitalized Covid-19 cases and 477,741 controls. This included cases reporting being on a ventilator, receiving oxygen, or having pneumonia compared to controls not reporting a positive test. In addition to the already reported locus on chromosome 3 (rs73064425, OR=2.14, discovery p=4.77×10-30), robust replication was found for new associations at four loci identified by GenOMICC: on chromosome 12 within the OAS gene cluster (rs74956615, OR=1.59, discovery p=1.65×10-8), near TYK2 on chromosome 19 (rs74956615, OR=1.59, discovery p=1.65×10-8), within DPP9 on chromosome 19 (rs2109069, OR=1.36, discovery p=3.98×10-12), and on chromosome 21 including the IFNAR2 gene (rs2236757, OR=1.28, discovery p=4.99×10-8) (Fig. 1, Extended Data Table 2).
To enhance the power of exploratory analyses, inverse-variance meta-analysis was conducted between GenOMICC severe EUR (ncases = 1676, ncontrols = 8380), UGI hospitalized Covid-19 vs. population without UK Biobank (B2, version 2) (ncases = 2415, ncontrols = 477741), and 23andMe broad respiratory phenotype (ncases = 1128, ncontrols = 679531). This revealed one additional genome-wide significant (but unreplicated) locus at CCHCR1 (considering no opportunity for replication in the meta-analysis, a stricter threshold of p<10-8 was used) (Table 2).
Mendelian Randomization
Mendelian randomization provides evidence for a causal relationship between an exposure variable and an outcome.18 We used two-sample summary data Mendelian randomization to evaluate evidence supporting the causal effects of RNA expression of various genes (GTEx v7, whole blood) on the odds of severe Covid-19. We identified a priori list of target genes related to the mechanisms of action of many host-targeted drugs proposed for treating Covid-19 (Supplementary Table 3). Among these targets, seven had suitable local expression quantitative trait loci (eQTL) in GTEx (v7). Of these, IFNAR2 remained significant after correcting for multiple testing of the seven tests by Bonferroni correction (β = -1.49, standard error = 0.52, p = 0.0043; Supplementary Table 4). There was uncertain evidence of heterogeneity (HEIDII19 p = 0.015), suggesting that this variant's effect on severe Covid-19 disease may be mediated through a different mechanism, leading to a potential over- or under-estimation of the effect of IFNAR2 expression on the risk of severe disease.
Next, we performed transcriptome-wide Mendelian randomization to quantify the support for non-selected genes as potential therapeutic targets. Instruments were available for 4,614 unique Ensembl gene IDs. After adjusting for multiple comparisons in this analysis, no gene was statistically significant (4,614 tests). After conservatively filtering for heterogeneity (HEIDI p > 0.05), the minimal Mendelian randomization p-value for the variant at chr19:10466123 influencing TYK2 expression was p = 0.00049. Nine other genes with nominally significant Mendelian randomization p-values (p < 0.0051) were advanced for further analysis (Supplementary Table 5).
To replicate these findings, we tested for external evidence using separate eQTL datasets (eQTLgen)20 and GWAS (HGI B2 excluding UK Biobank). Mendelian randomization signals with tentative directional effects were significant for IFNAR2 (p = 7.5×10-4) and TYK2 (p = 5.5×10-5; Supplementary Table 6).
Transcriptome-Wide Association Study
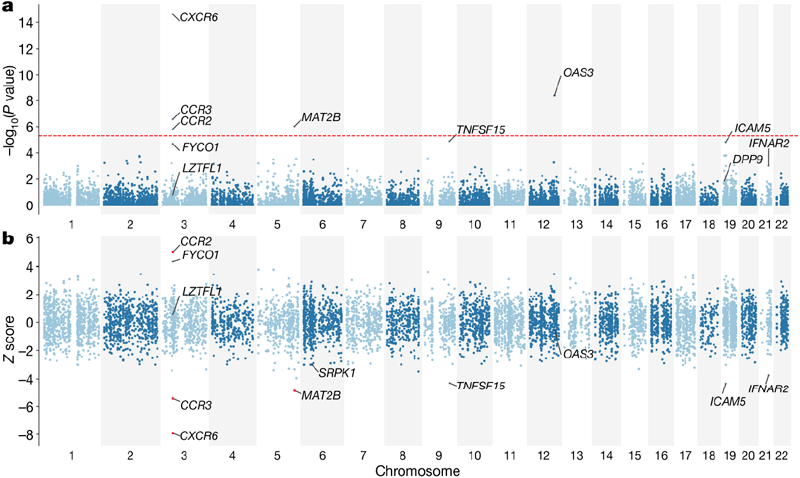
By inferring gene expression from known genetic variants associated with transcript levels (eQTLs), we performed a transcriptome-wide association study (TWAS)21,22 to link GWAS results with tissue-specific gene expression data. This analysis utilized GTEx v8 data from two pre-selected disease-relevant tissues (whole blood and lung) (Figure 2). We selected genes with p < 0.05 in these tissues and incorporated eQTL data from other tissues in GTEx into a composite meta-TWAS analysis23 to optimize the power to detect differences in predicted expression in lung or blood. We identified five genes with genome-wide significant differences in predicted expression (Supplementary Table 7). Among these were four genes with differences in predicted expression in lung tissue (three genes on chr3). We then compared these results with existing biological knowledge about host-virus interactions in Covid-19 using information-based meta-analysis (MAIC)24. The GenOMICC TWAS results overlapped more with transcriptome, proteome, and CRISPR studies of host genes involved in Covid-19 than any other data source (Extended Data Figure 2).
Genetic Correlation
Using the high-definition likelihood (HDL) method25, we initially estimated the SNP-based heritability (the proportion of phenotypic variance captured by additive effects at common SNPs) of severe Covid-19 at 0.065 (SE = 0.019). Two additional analyses did not detect significant signals. The first analysis used controls from the 100,000 Genomes Project, where matching with GenOMICC cases was less close, potentially limiting heritability estimates. The second analysis involved a smaller GWAS comparing some GenOMICC cases to controls from the UK Biobank, matched as closely as possible for BMI and age. This second analysis was less powerful due to the limited number of tightly matched cases (ncases = 1260; ncontrols = 6300; Supplementary Figure 14). Future analyses incorporating a larger number of cases and rare variants should provide more comprehensive heritability estimates. We also examined genetic correlations with other traits, i.e., the extent to which underlying genetic components are shared with severe Covid-19. Using the HDL method, we identified significant negative genetic correlations with educational attainment and intelligence. Additionally, significant positive genetic correlations were detected with several adiposity phenotypes, such as body mass index and fat mass in the legs (Supplementary Figure 19). Furthermore, as seen in GWAS results for other infectious and inflammatory diseases, variants strongly associated with promoters and enhancers were significantly enriched (Supplementary Figure 18).
Discussion
We discovered and replicated significant genetic associations with life-threatening Covid-19 (Figure 1). By focusing on severe cases, some of these associations likely relate to late-stage immune-mediated diseases, such as respiratory failure requiring invasive mechanical ventilation2. Crucially, the genome-wide unbiased nature of GWAS allows for the discovery of entirely novel pathophysiological mechanisms. Genetic variation can be used to infer causality, providing genetic evidence supporting therapeutic targets, thereby substantially increasing the likelihood of successful drug development. Specifically, Mendelian randomization occupies a unique position in the hierarchy of clinical evidence.
Patients admitted to UK intensive care units during the first wave of Covid-19 were, on average, younger and had fewer comorbidities. The cohort studied here is defined by a tendency towards severe respiratory failure caused by Covid-19. In GenOMICC, we recruited from 208 intensive care units (covering 95% of UK ICU capacity) to ensure broad genetic ancestry coverage of UK patients (Extended Data Figure 3).
For external replication, the closest comparisons are the analyses of hospitalized patients and population controls from the Covid-19 Host Genetics Initiative and the broad respiratory phenotype from 23andMe, both generously shared with the international community. Likewise, the complete summary statistics from GenOMICC are immediately available at genomicc.org/data.
Despite differences in case definitions, novel associations identified in studies of severe cases were robustly replicated in combined data from hospitalization studies (Extended Data Table 2). Moreover, Mendelian randomization results suggesting causality for IFNAR2 and TYK2 were statistically significant in confirmatory analyses. Our findings reveal that severe Covid-19 is associated with at least two biological mechanisms: innate antiviral defenses (IFNAR2 and OAS genes) known to be crucial early in disease and host-driven inflammatory lung injury (DPP9, TYK2, and CCR2) central to life-threatening late Covid-19.
Interferons are host antiviral signaling mediators that stimulate the release of many essential components of the early host response to viral infection30. Consistent with the beneficial role of type I interferons, increased expression of the interferon receptor subunit IFNAR2 reduced the odds of severe Covid-19 (Mendelian randomization discovery p = 0.0043 [7 tests]; replication p = 7.5×10-4 [1 test]). Within the assumptions of Mendelian randomization, this represents evidence of a protective role for IFNAR2 in Covid-19. Rare loss-of-function variants in IFNAR2 are associated with severe Covid-1912 and many other viral diseases31,32. This suggests that interferon administration may reduce the likelihood of severe Covid-19, though our evidence cannot distinguish the disease phase where such therapy would be effective. It implies that the genetic effect may be mediated during the early stages of the disease when viral load is high33.
The variant rs10735079 (chr12, p = 1.65 × 10-8) is located in the cluster of interferon-induced oligoadenylate synthetase (OAS) genes (OAS1, OAS2, OAS3; Figure 1). Our TWAS detected a significant association with predicted expression of OAS3 (Figure 2). OAS1 variants have been implicated in SARS-CoV susceptibility in candidate gene association studies conducted in Vietnam34 and China35. These genes encode enzymes that produce the mediator (2',5'-oligoadenylate 2-5A) activating the effector enzyme RNAse L, which degrades double-stranded RNA, a replication intermediate of coronaviruses36. Betacoronaviruses OC43 and MHV produce phosphodiesterases that cleave the host antiviral mediator 2-5A38, but SARS-CoV-2 is not known to possess this capability. OAS genes are potential therapeutic targets since endogenous phosphodiesterase 12 (PDE-12) activity degrades the host antiviral mediator 2-5A39. PDE-12 inhibitors that enhance antiviral activity mediated through OAS are available.
The association at 19p13.3 (rs2109069, p = 3.98 × 10-12) is an intronic variant in the gene encoding dipeptidyl peptidase 9 (DPP9). Variants in this locus are associated with idiopathic pulmonary fibrosis40. DPP9 encodes a serine protease with diverse intracellular functions, including the cleavage of the antiviral signaling molecule CXCL1041, antigen presentation, and activation of inflammatory cells42. Identifying genes that cause inflammatory organ damage is crucial. TYK2 is one of the four genetic targets of JAK inhibitors like baricitinib44, one of the candidate drugs used in our a priori target list (Supplementary Table 3). The association between TYK2 expression and disease severity is confirmed in external datasets.
We reproduce the findings of Ellinghaus et al. at 3p21.31.8. The very small p-values at this locus (p=4.77×10-30) may reflect the larger scale of our study and the focus on extreme severity. Many genes at this locus could potentially explain the association. Systematic review and meta-analysis of experimental data on betacoronavirus infection provided biological support for FYCO145. Our TWAS results indicate that variants in this region significantly affect predicted expression of CXCR6, CCR2, and CCR3 (Figure 2a). One or more of these genes likely mediates the critical pathophysiological mechanism in severe disease.
The association between predicted expression of CCR2 (CC-chemokine receptor 2) and severe disease is particularly strong in lung tissue (Figure 2b). CCR2 promotes monocyte/macrophage chemotaxis to inflammation sites, and its major ligand (monocyte chemoattractant protein/MCP-1) is upregulated in bronchoalveolar lavage fluid from mechanically ventilated Covid-19 patients46. Circulating MCP-1 levels are associated with more severe disease. Anti-CCR2 monoclonal antibody therapy is safe in rheumatoid arthritis47. While the ABO locus has been previously associated with Covid-198, it was not genome-wide significant in our GenOMICC severe cohort48. Interestingly, the combined meta-analysis showed a near genome-wide significant signal at this locus, suggesting that this variant might influence susceptibility to Covid-19 rather than severity (Extended Data Figure 4).
Analysis of shared heritability revealed positive correlations with adiposity traits. This does not suggest causality due to potential biases but likely reflects a combination of two effects. First, increased BMI and lower socioeconomic status are strong risk factors for severe Covid-1914, and second, UK Biobank participants are disproportionately drawn from societal groups underrepresented in obesity49.
Completing and reporting this study urgently necessitated using controls genotyped using different techniques than the cases, drawn from population genetic studies with systematic differences in population structure, demographics, and comorbidities. Residual confounding is reflected in the lambda (λ0.5) values of 1.099 in our primary analysis (Extended Data Figure 1). We mitigated the risk of false-positive associations due to genotyping errors by genotyping the majority of subjects using two different methods (microarray and whole-genome sequencing) and validating significant associations with two additional control groups (100,000 Genomes and Generation Scotland). The success of these mitigations is demonstrated by the robust replication of our sentinel SNPs in external studies. Our meta-analysis combining GenOMICC and multiple additional sources of genome-wide association yielded an encouraging λ0.5 = 1.017 (Extended Data Figure 1).
Further studies to deepen these findings are imperative. Our MAIC results indicate that genes ranked highly in GenOMICC are likely involved in Covid-19 in other studies (Extended Data Figure 2). We continue recruiting into the GenOMICC study, expecting additional associations to be present and detectable with larger case numbers. Future studies using whole-genome sequencing will explore the rarer end of the allele frequency spectrum for variants that increase susceptibility. Given that the GenOMICC cohort is strongly enriched for immediately life-threatening disease, defined by patients receiving invasive mechanical ventilation or judged by the treating clinician as being at high risk of requiring mechanical support, effect sizes are likely to be larger.
We discovered novel genetic associations with severe Covid-19. Some of these associations directly lead to potential therapeutic approaches, such as augmenting interferon signaling, antagonizing monocyte activation and infiltration into the lung, or specifically targeting detrimental inflammatory pathways. This significantly contributes to the biological rationale supporting specific treatments, but each must be tested in large clinical trials before incorporation into clinical practice.
Online Content
Methods, Additional References, Nature Research Reporting Summary, Source Data, Extended Data, Supplementary Information, Acknowledgements, Peer Review Information, Author Contributions and Competing Interests, Data and Code Availability statements are available at https://doi.org/10.1038/s41586-020-03065-y.
- Dorward, D.A., Russell, C.D., Um, I.H., Elshani, M., Armstrong, S.D., Penrice-Randal, R., Millar, T., Lerpiniere, C.E., Tagliavini, G., Hartley, C.S., Randle, N.P., Gachanja, N.N., Potey, P.M., Dong, X., Anderson, A.M., Campbell, V.L., Duguid, A.J., Al Qsous, W., BouHaidar, R., Baillie, J.K., Dhaliwal, K., Wallace, W.A., Bellamy, C.O., Prost, S., Smith, C., Hiscox, J.A., Harrison, D.J. & Lucas, C.D. Tissue-specific immune pathology in fatal Covid-19. American Journal of Respiratory and Critical Care Medicine (2020). https://doi.org/10.1164/rccm.202008-3265OC
- Horby, P., Lim, W.S., Emberson, J.R., Mafham, M., Bell, J.L., Linsell, L., Staplin, N., Brightling, C., Ustianowski, A., Elmahi, E., Prudon, B., Green, C., Felton, T., Chadwick, D., Rege, K., Fegan, C., Chappell, L.C., Faust, S.N., Jaki, T., Jeffery, K., Montgomery, A., Rowan, K., Juszczak, E., Baillie, J.K., Haynes, R. & Landray, M.J. Dexamethasone in hospitalized patients with Covid-19 - Preliminary report. New England Journal of Medicine (2020). https://doi.org/10.1056/NEJMoa2021436
- Baillie, J.K. Translational genomics targeting host immune responses to fight infections. Science (New York, N.Y.) 344, 807–8 (2014).
- Sørensen, T.I., Nielsen, G.G., Andersen, P.K. & Teasdale, T.W. Genetic and environmental influences on premature death in adult adoptees. New England Journal of Medicine 318, 727–32 (1988).
- Patarčić, I., Gelemanović, A., Kirin, M., Kolčić, I., Theodoratou, E., Baillie, J.K., de Jong, M.D., Rudan, I., Campbell, H. & Polašek, O. Role of host genetic factors in respiratory infections: a systematic review, meta-analysis, and field synopsis. Scientific Reports 5, 16119 (2015).
- Horby, P., Nguyen, N.Y., Dunstan, S.J. & Baillie, J.K. The role of host genetics in susceptibility to influenza: a systematic review. Influenza and Other Respiratory Viruses 7 2, 37–41 (2013).
- Clohisey, S. & Baillie, J.K. Host susceptibility to severe influenza A virus infection. Critical Care 23, 303(2019).
- Ellinghaus, D., Degenhardt, F., Bujanda, L., Buti, M., Albillos, A., Invernizzi, P., Fernández, J., Prati, D., Baselli, G., Asselta, R., Grimsrud, M.M., Milani, C., Aziz, F., Kässens, J., May, S., Wendorff, M., Wienbrandt, L., Uellendahl-Werth, F., Zheng, T., Yi, X., de Pablo, R., Chercoles, A.G., Palom, A., Garcia-Fernandez, A.-E., Rodriguez-Frias, F., Zanella, A., Bandera, A., Protti, A., Aghemo, A., Lleo, A., Biondi, A., Caballero-Garralda, A., Gori, A., Tanck, A., Carreras Nolla, A., Latiano, A., Fracanzani, A.L., Peschuck, A., Julià, A., Pesenti, A., Voza, A., Jiménez, D., Mateos, B., Nafria Jimenez, B., Quereda, C., Paccapelo, C., Gassner, C., Angelini, C., Cea, C., Solier, A., Pestaña, D., Muñiz-Diaz, E., Sandoval, E., Paraboschi, E.M., Navas, E., Garcı́a Sánchez, F., Ceriotti, F., Martinelli-Boneschi, F., Peyvandi, F., Blasi, F., Téllez, L., Blanco-Grau, A., Hemmrich-Stanisak, G., Grasselli, G., Costantino, G., Cardamone, G., Foti, G., Aneli, S., Kurihara, H., ElAbd, H., My, I., Galván-Femenia, I., Martı́n, J., Erdmann, J., Ferrusquı́a-Acosta, J., Garcia-Etxebarria, K., Izquierdo-Sanchez, L., Bettini, L.R., Sumoy, L., Terranova, L., Moreira, L., Santoro, L., Scudeller, L., Mesonero, F., Roade, L., Rühlemann, M.C., Schaefer, M., Carrabba, M., Riveiro-Barciela, M., Figuera Basso, M.E., Valsecchi, M.G., Hernandez-Tejero, M., Acosta-Herrera, M., DAngiò, M., Baldini, M., Cazzaniga, M., Schulzky, M., Cecconi, M., Wittig, M., Ciccarelli, M., Rodrı́guez-Gandı́a, M., Bocciolone, M., Miozzo, M., Montano, N., Braun, N., Sacchi, N., Martı́nez, N., Özer, O., Palmieri, O., Faverio, P., Preatoni, P., Bonfanti, P., Omodei, P., Tentorio, P., Castro, P., Rodrigues, P.M., Blandino Ortiz, A., Cid, R. de, Ferrer, R., Gualtierotti, R., Nieto, R., Goerg, S., Badalamenti, S., Marsal, S., Matullo, G., Pelusi, S., Juzenas, S., Aliberti, S., Monzani, V., Moreno, V., Wesse, T., Lenz, T.L., Pumarola, T., Rimoldi, V., Bosari, S., Albrecht, W., Peter, W., Romero-Gómez, M., DAmato, M., Duga, S., Banales, J.M., Hov, J.R., Folseraas, T., Valenti, L., Franke, A. & Karlsen, T.H. Genome-wide association study of severe Covid-19 with respiratory failure. New England Journal of Medicine (2020). https://doi.org/10.1056/NEJMoa2020283
- Shelton, J.F., Shastri, A.J., Ye, C., Weldon, C.H., Filshtein-Somnez, T., Coker, D., Symons, A., Esparza-Gordillo, J., Team, T.2.C.-1., Aslibekyan, S. & Auton, A. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. medRxiv 2020.09.04.20188318 (2020). https://doi.org/10.1101/2020.09.04.20188318
- Casanova, J.-L. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proceedings of the National Academy of Sciences of the United States of America 112, E7128–37 (2015).
- Plenge, R.M. Molecular basis of severe coronavirus disease 2019. JAMA (2020). https://doi.org/10.1001/jama.2020.14015
- Zhang, Q., Bastard, P., Liu, Z., Pen, J.L., Moncada-Velez, M., Chen, J., Ogishi, M., Sabli, I.K.D., Hodeib, S., Korol, C., Rosain, J., Bilguvar, K., Ye, J., Bolze, A., Bigio, B., Yang, R., Arias, A.A., Zhou, Q., Zhang, Y., Onodi, F., Korniotis, S., Karpf, L., Philippot, Q., Chbihi, M., Bonnet-Madin, L., Dorgham, K., Smith, N., Schneider, W.M., Razooky, B.S., Hoffmann, H.-H., Michailidis, E., Moens, L., Han, J.E., Lorenzo, L., Bizien, L., Meade, P., Neehus, A.-L., Ugurbil, A.C., Corneau, A., Kerner, G., Zhang, P., Rapaport, F., Seeleuthner, Y., Manry, J., Masson, C., Schmitt, Y., Schlüter, A., Voyer, T.L., Khan, T., Li, J., Fellay, J., Roussel, L., Shahrooei, M., Alosaimi, M.F., Mansouri, D., Al-Saud, H., Al-Mulla, F., Almourfi, F., Al-Muhsen, S.Z., Alsohime, F., Turki, S.A., Hasanato, R., van de Beek, D., Biondi, A., Bettini, L.R., D’Angio, M., Bonfanti, P., Imberti, L., Sottini, A., Paghera, S., Quiros-Roldan, E., Rossi, C., Oler, A.J., Tompkins, M.F., Alba, C., Vandernoot, I., Goffard, J.-C., Smits, G., Migeotte, I., Haerynck, F., Soler-Palacin, P., Martin-Nalda, A., Colobran, R., Morange, P.-E., Keles, S., Çölkesen, F., Ozcelik, T., Yasar, K.K., Senoglu, S., Karabela, Ş.N., Gallego, C.R., Novelli, G., Hraiech, S., Tandjaoui-Lambiotte, Y., Duval, X., Laouénan, C., C-S, Clinicians, C., I.C, Group, F.C.C.S., C-C, Cohort, A.U, Covid-19, Biobank, C.H.G, Effort, Niaid-Usuhs, Group, T.C.I, Snow, A.L., Dalgard, C.L., Milner, J., Vinh, D.C., Mogensen, T.H., Marr, N., Spaan, A.N., Boisson, B., Boisson-Dupuis, S., Bustamante, J., Puel, A., Ciancanelli, M., Meyts, I., Maniatis, T., Soumelis, V., Amara, A., Nussenzweig, M., García-Sastre, A., Krammer, F., Pujol, A., Duffy, D., Lifton, R., Zhang, S.-Y., Gorochov, G., Béziat, V., Jouanguy, E., Sancho-Shimizu, V., Rice, C.M., Abel, L., Notarangelo, L.D., Cobat, A., Su, H.C. & Casanova, J.-L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science (2020). https://doi.org/10.1126/science.abd4570
- Millar, J.E., Neyton, L., Seth, S., Dunning, J., Merson, L., Murthy, S., Russell, C.D., Keating, S., Swets, M., Sudre, C.H., Spector, T.D., Ourselin, S., Steves, C.J., Wolf, J., Investigators, I., Docherty, A.B., Harrison, E.M., Openshaw, P.J., Semple, M.G. & Baillie, J.K. Robust and reproducible clinical patterns in hospitalised patients with COVID-19. medRxiv 2020.08.14.20168088 (2020). https://doi.org/10.1101/2020.08.14.20168088
- Docherty, A.B., Harrison, E.M., Green, C.A., Hardwick, H.E., Pius, R., Norman, L., Holden, K.A., Read, J.M., Dondelinger, F., Carson, G., Merson, L., Lee, J., Plotkin, D., Sigfrid, L., Halpin, S., Jackson, C., Gamble, C., Horby, P.W., Nguyen-Van-Tam, J.S., Ho, A., Russell, C.D., Dunning, J., Openshaw, P.J., Baillie, J.K. & Semple, M.G. Features of 200.167em133 UK patients hospitalised with Covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ (Clinical Research Ed.). 369, m1985 (2020).
- Angus, D.C., Derde, L., Al-Beidh, F., Annane, D., Arabi, Y., Beane, A., van Bentum-Puijk, W., Berry, L., Bhimani, Z., Bonten, M., Bradbury, C., Brunkhorst, F., Buxton, M., Buzgau, A., Cheng, A.C., de Jong, M., Detry, M., Estcourt, L., Fitzgerald, M., Goossens, H., Green, C., Haniffa, R., Higgins, A.M., Horvat, C., Hullegie, S.J., Kruger, P., Lamontagne, F., Lawler, P.R., Linstrum, K., Litton, E., Lorenzi, E., Marshall, J., McAuley, D., McGlothin, A., McGuinness, S., McVerry, B., Montgomery, S., Mouncey, P., Murthy, S., Nichol, A., Parke, R., Parker, J., Rowan, K., Sanil, A., Santos, M., Saunders, C., Seymour, C., Turner, A., van de Veerdonk, F., Venkatesh, B., Zarychanski, R., Berry, S., Lewis, R.J., McArthur, C., Webb, S.A. & Gordon, A.C. Effect of hydrocortisone on mortality and organ support in patients with severe COVID-19: the REMAP-CAP COVID-19 Corticosteroid Domain Randomized Clinical Trial. JAMA (2020). https://doi.org/10.1001/jama.2020.17022
- Carvelli, J., Demaria, O., Vély, F., Batista, L., Benmansour, N.C., Fares, J., Carpentier, S., Thibult, M.-L., Morel, A., Remark, R., André, P., Represa, A., Piperoglou, C., Cordier, P.Y., Le Dault, E., Guervilly, C., Simeone, P., Gainnier, M., Morel, Y., Ebbo, M., Schleinitz, N. & Vivier, E. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature (2020). https://doi.org/10.1038/s41586-020-2600-6
- Baillie, J.K. & Digard, P. Influenza Time to Target the Host? New England Journal of Medicine 369, 191–193(2013).
- Bretherick, A.D., Canela-Xandri, O., Joshi, P.K., Clark, D.W., Rawlik, K., Boutin, T.S., Zeng, Y., Amador, C., Navarro, P., Rudan, I., Wright, A.F., Campbell, H., Vitart, V., Hayward, C., Wilson, J.F., Tenesa, A., Ponting, C.P., Baillie, J.K. & Haley, C. Linking protein to phenotype with Mendelian randomization detects 38 proteins with causal roles in human diseases and traits. PLoS Genetics 16, e1008785 (2020).
- Zhu, Z., Zhang, F., Hu, H., Bakshi, A., Robinson, M.R., Powell, J.E., Montgomery, G.W., Goddard, M.E., Wray, N.R., Visscher, P.M. & Yang, J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genetics 48, 481–7 (2016).
- Võsa, U., Claringbould, A., Westra, H.-J., Bonder, M.J., Deelen, P., Zeng, B., Kirsten, H., Saha, A., Kreuzhuber, R., Kasela, S., Pervjakova, N., Alvaes, I., Fave, M.-J., Agbessi, M., Christiansen, M., Jansen, R., Seppälä, I., Tong, L., Teumer, A., Schramm, K., Hemani, G., Verlouw, J., Yaghootkar, H., Sönmez, R., Brown, A., Kukushkina, V., Kalnapenkis, A., Rüeger, S., Porcu, E., Kronberg-Guzman, J., Kettunen, J., Powell, J., Lee, B., Zhang, F., Arindrarto, W., Beutner, F., Consortium, B., Brugge, H., Consortium, i., Dmitreva, J., Elansary, M., Fairfax, B.P., Georges, M., Heijmans, B.T., Kähönen, M., Kim, Y., Knight, J.C., Kovacs, P., Krohn, K., Li, S., Loeffler, M., Marigorta, U.M., Mei, H., Momozawa, Y., Müller-Nurasyid, M., Nauck, M., Nivard, M., Penninx, B., Pritchard, J., Raitakari, O., Rotzchke, O., Slagboom, E.P., Stehouwer, C.D.A., Stumvoll, M., Sullivan, P., Hoen, P.A.C.‘., Thiery, J., Tönjes, A., Dongen, J. van, Iterson, M. van, Veldink, J., Völker, U., Wijmenga, C., Swertz, M., Andiappan, A., Montgomery, G.W., Ripatti, S., Perola, M., Kutalik, Z., Dermitzakis, E., Bergmann, S., Frayling, T., Meurs, J. van, Prokisch, H., Ahsan, H., Pierce, B., Lehtimäki, T., Boomsma, D., Psaty, B.M., Gharib, S.A., Awadalla, P., Milani, L., Ouwehand, W., Downes, K., Stegle, O., Battle, A., Yang, J., Visscher, P.M., Scholz, M., Gibson, G., Esko, T. & Franke, L. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv 447367 (2018). https://doi.org/10.1101/447367
- Gamazon, E.R., Wheeler, H.E., Shah, K.P., Mozaffari, S.V., Aquino-Michaels, K., Carroll, R.J., Eyler, A.E., Denny, J.C., Nicolae, D.L., Cox, N.J. & Im, H.K. A gene-based association method for mapping traits using reference transcriptome data. Nature Genetics 47, 1091–8 (2015).
- Gusev, A., Ko, A., Shi, H., Bhatia, G., Chung, W., Penninx, B.W.J.H., Jansen, R., de Geus, E.J.C., Boomsma, D.I., Wright, F.A., Sullivan, P.F., Nikkola, E., Alvarez, M., Civelek, M., Lusis, A.J., Lehtimäki, T., Raitoharju, E., Kähönen, M., Seppälä, I., Raitakari, O.T., Kuusisto, J., Laakso, M., Price, A.L., Pajukanta, P. & Pasaniuc, B. Integrative approaches for large-scale transcriptome-wide association studies. Nature Genetics 48, 245–52 (2016).
- Barbeira, A.N., Dickinson, S.P., Bonazzola, R., Zheng, J., Wheeler, H.E., Torres, J.M., Torstenson, E.S., Shah, K.P., Garcia, T., Edwards, T.L., Stahl, E.A., Huckins, L.M., Nicolae, D.L., Cox, N.J. & Im, H.K. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nature Communications 9, 1825 (2018).
- Li, B., Clohisey, S.M., Chia, B.S., Wang, B., Cui, A., Eisenhaure, T., Schweitzer, L.D., Hoover, P., Parkinson, N.J., Nachshon, A., Smith, N., Regan, T., Farr, D., Gutmann, M.U., Bukhari, S.I., Law, A., Sangesland, M., Gat-Viks, I., Digard, P., Vasudevan, S., Lingwood, D., Dockrell, D.H., Doench, J.G., Baillie, J.K. & Hacohen, N. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nature Communications 11, 164 (2020).
- Ning, Z., Pawitan, Y. & Shen, X. High-definition likelihood inference of genetic correlations across human complex traits. Nature Genetics 52, 859–864 (2020).
- Forrest, A.R.R., Kawaji, H., Rehli, M., Baillie, J.K., et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
- Villar, D., Berthelot, C., Aldridge, S., Rayner, T.F., Lukk, M., Pignatelli, M., Park, T.J., Deaville, R., Erichsen, J.T., Jasinska, A.J., Turner, J.M.A., Bertelsen, M.F., Murchison, E.P., Flicek, P. & Odom, D.T. Enhancer evolution across 20 mammalian species. Cell 160, 554–66 (2015).
- Plenge, R.M., Scolnick, E.M. & Altshuler, D. Validating therapeutic targets through human genetics. Nature Reviews Drug Discovery 12, 581–594 (2013).
- Davies, N.M., Holmes, M.V. & Smith, G.D. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601 (2018).
- Sadler, A.J. & Williams, B.R.G. Interferon-inducible antiviral effectors. Nature Reviews. Immunology 8, 559–68 (2008).
- Hambleton, S., Goodbourn, S., Young, D.F., Dickinson, P., Mohamad, S.M.B., Valappil, M., McGovern, N., Cant, A.J., Hackett, S.J., Ghazal, P., Morgan, N.V. & Randall, R.E. STAT2 deficiency and susceptibility to viral illness in humans. Proceedings of the National Academy of Sciences of the United States of America 110, 3053–8 (2013).
- Duncan, C.J.A., Mohamad, S.M.B., Young, D.F., Skelton, A.J., Leahy, T.R., Munday, D.C., Butler, K.M., Morfopoulou, S., Brown, J.R., Hubank, M., Connell, J., Gavin, P.J., McMahon, C., Dempsey, E., Lynch, N.E., Jacques, T.S., Valappil, M., Cant, A.J., Breuer, J., Engelhardt, K.R., Randall, R.E. & Hambleton, S. Human IFNAR2 deficiency: lessons for antiviral immunity. Science Translational Medicine 7, 307ra154 (2015).
- Consortium, W.S. trialPan, H., Peto, R., Karim, Q.A., Alejandria, M., Henao-Restrepo, A.M., García, C.H., Kieny, M.-P., Malekzadeh, R., Murthy, S., Preziosi, M.-P., Reddy, S., Periago, M.R., Sathiyamoorthy, V., Røttingen, J.-A., Swaminathan, S. & Writing Committee Members, A.R., T.A., C., I. Repurposed antiviral drugs for COVID-19—interim WHO SOLIDARITY trial results. medRxiv 2020.10.15.20209817 (2020). https://doi.org/10.1101/2020.10.15.20209817
- Hamano, E., Hijikata, M., Itoyama, S., Quy, T., Phi, N.C., Long, H.T., Ha, L.D., Ban, V.V., Matsushita, I., Yanai, H., Kirikae, F., Kirikae, T., Kuratsuji, T., Sasazuki, T. & Keicho, N. Polymorphisms of interferon-inducible genes OAS-1 and MXA in a Vietnamese population: relation to severe acute respiratory syndrome. Biochemical and Biophysical Research Communications 329, 1234–9 (2005).
- He, J., Feng, D., de Vlas, S.J., Wang, H., Fontanet, A., Zhang, P., Plancoulaine, S., Tang, F., Zhan, L., Yang, H., Wang, T., Richardus, J.H., Habbema, J.D.F. & Cao, W. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MXA genes: a case-control study. BMC Infectious Diseases 6, 106 (2006).
- Choi, U.Y., Kang, J.-S., Hwang, Y.S. & Kim, Y.-J. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Experimental & Molecular Medicine 47, e144 (2015).
- Hagemeijer, M.C., Vonk, A.M., Monastyrska, I., Rottier, P.J.M. & Haan, C.A.M. de Visualizing coronavirus rna synthesis in time by using click chemistry. Journal of virology 86, 5808–16 (2012).
- Silverman, R.H. & Weiss, S.R. Viral phosphodiesterases antagonize RNA silencing by degrading 2-5A to repress RNase L signaling. Journal of Interferon & Cytokine Research 34, 455–463 (2014).
- Wood, E.R., Bledsoe, R., Chai, J., Daka, P., Deng, H., Ding, Y., Harris-Gurley, S., Kryn, L.H., Nartey, E., Nichols, J., Nolte, R.T., Prabhu, N., Rise, C., Sheahan, T., Shotwell, J.B., Smith, D., Tai, V., Taylor, J.D., Tomberlin, G., Wang, L., Wisely, B., You, S., Xia, B. & Dickson, H. Role of phosphodiesterase 12 (PDE12) as a negative regulator of innate immunity and its identification as an antiviral target. Journal of Biological Chemistry jbc.M115.653113 (2015). https://doi.org/10.1074/jbc.M115.653113
- Fingerlin, T.E., Murphy, E., Zhang, W., Peljto, A.L., Brown, K.K., Steele, M.P., Loyd, J.E., Cosgrove, G.P., Lynch, D., Groshong, S., Collard, H.R., Wolters, P.J., Bradford, W.Z., Kossen, K., Seiwert, S.D., du Bois, R.M., Garcia, C.K., Devine, M.S., Gudmundsson, G., Isaksson, H.J., Kaminski, N., Zhang, Y., Gibson, K.F., Lancaster, L.H., Cogan, J.D., Mason, W.R., Maher, T.M., Molyneaux, P.L., Wells, A.U., Moffatt, M.F., Selman, M., Pardo, A., Kim, D.S., Crapo, J.D., Make, B.J., Regan, E.A., Walek, D.S., Daniel, J.J., Kamatani, Y., Zelenika, D., Smith, K., McKean, D., Pedersen, B.S., Talbert, J., Kidd, R.N., Markin, C.R., Beckman, K.B., Lathrop, M., Schwarz, M.I. & Schwartz, D.A. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nature Genetics 45, 613–20 (2013).
- Zhang, H., Maqsudi, S., Rainczuk, A., Duffield, N., Lawrence, J., Keane, F.M., Justa-Schuch, D., Geiss-Friedlander, R., Gorrell, M.D. & Stephens, A.N. Identification of novel dipeptidyl peptidase 9 substrates by two-dimensional differential in-gel electrophoresis. The FEBS Journal 282, 3737–57 (2015).
- Geiss-Friedlander, R., Parmentier, N., Möller, U., Urlaub, H., Van den Eynde, B.J. & Melchior, F. The cytoplasmic peptidase DPP9 is a rate-limiting factor for degradation of proline-containing peptides. The Journal of Biological Chemistry 284, 27211–9 (2009).
- Griswold, A.R., Ball, D.P., Bhattacharjee, A., Chui, A.J., Rao, S.D., Taabazuing, C.Y. & Bachovchin, D.A. DPP9's enzyme activity inhibits inflammasome activation by a mechanism distinct from its binding to CARD8. ACS Chemical Biology 14, 2424–2429 (2019).
- Nguyen, D.-T., Mathias, S., Bologa, C., Brunak, S., Fernandez, N., Gaulton, A., Hersey, A., Holmes, J., Jensen, L.J., Karlsson, A., Liu, G., Maayan, A., Mandava, G., Mani, S., Mehta, S., Overington, J., Patel, J., Rouillard, A.D., Schürer, S., Sheils, T., Simeonov, A., Sklar, L.A., Southall, N., Ursu, O., Vidovic, D., Waller, A., Yang, J., Jadhav, A., Oprea, T.I. & Guha, R. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Research 45, D995–D1002 (2017).
- Parkinson, N., Rodgers, N., Fourman, M.H., Wang, B., Zechner, M., Swets, M., Millar, J.E., Law, A., Russell, C., Baillie, J.K. & Clohisey, S. Systematic review and meta-analysis identifies potential host therapeutic targets for COVID-19. medRxiv 2020.08.27.20182238 (2020). https://doi.org/10.1101/2020.08.27.20182238
- Zhou, Z., Ren, L., Zhang, L., Zhong, J., Xiao, Y., Jia, Z., Guo, L., Yang, J., Wang, C., Jiang, S., Yang, D., Zhang, G., Li, H., Chen, F., Xu, Y., Chen, M., Gao, Z., Yang, J., Dong, J., Liu, B., Zhang, X., Wang, W., He, K., Jin, Q., Li, M. & Wang, J. Heightened innate immune responses in the respiratory tract of covid-19 patients. Cell host & microbe 27, 883–890.e2 (2020).
- Zhao, Y., Qin, L., Zhang, P., Li, K., Liang, L., Sun, J., Xu, B., Dai, Y., Li, X., Zhang, C., Peng, Y., Feng, Y., Li, A., Hu, Z., Xiang, H., Ogg, G., Ho, L.-P., McMichael, A., Jin, R., Knight, J.C., Dong, T. & Zhang, Y. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight 5, (2020).
- Vergunst, C.E., Gerlag, D.M., Lopatinskaya, L., Klareskog, L., Smith, M.D., van den Bosch, F., Dinant, H.J., Lee, Y., Wyant, T., Jacobson, E.W., Baeten, D. & Tak, P.P. Modulation of CCR2 in rheumatoid arthritis: a double-blind, randomized, placebo-controlled clinical trial. Arthritis and Rheumatism 58, 1931–9 (2008).
- Sudlow, C., Gallacher, J., Allen, N., Beral, V., Burton, P., Danesh, J., Downey, P., Elliott, P., Green, J., Landray, M., Liu, B., Matthews, P., Ong, G., Pell, J., Silman, A., Young, A., Sprosen, T., Peakman, T. & Collins, R. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Medicine 12, e1001779 (2015).
Publisher’s Note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
© The Author(s), published under an exclusive license to Springer Nature Limited 2020.
1Roslin Institute, University of Edinburgh, Easter Bush, Edinburgh, EH25 9RG, UK. 2MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Western General Hospital, Crewe Road, Edinburgh, EH4 2XU, UK. 3Genomics England, London, UK. 4Centre for Inflammation Research, The Queen’s Medical Research Institute, University of Edinburgh, 47 Little France Crescent, Edinburgh, UK. 5Centre for Genomic and Experimental Medicine, Institute of Genetics and Molecular Medicine, University of Edinburgh, Western General Hospital, Crewe Road, Edinburgh, EH4 2XU, UK. 6Edinburgh Clinical Research Facility, Western General Hospital, University of Edinburgh, Edinburgh, EH4 2XU, UK. 7Intensive Care National Audit & Research Centre, London, UK. 8Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland, Australia. 9Biostatistics Group, School of Life Sciences, Sun Yat-sen University, Guangzhou, China. 10The Crick Institute, London, UK. 11Intensive Care Unit, Royal Infirmary of Edinburgh, 54 Little France Drive, Edinburgh, EH16 5SA, UK. 12School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China. 13Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China. 14Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK. 15Department of Medicine, University of Cambridge, Cambridge, UK. 16Department of Intensive Care Medicine, Guy’s and St. Thomas NHS Foundation Trust, London, UK. 17School of Immunology and Microbial Sciences, King’s College London, London, UK. 18NIHR Health Protection Research Unit for Emerging and Zoonotic Infections, Institute of Infection, Veterinary and Ecological Sciences University of Liverpool, Liverpool, L69 7BE, UK. 19MRC-University of Glasgow Centre for Virus Research, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, UK. 20William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, EC1M 6BQ, UK. 21Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Old Road Campus, Roosevelt Drive, Oxford, OX3 7FZ, UK. 22Clinical Research Centre at St Vincent’s University Hospital, University College Dublin, Dublin, Ireland. 23Australian and New Zealand Intensive Care Research Centre, Monash University, Melbourne, Victoria, Australia. 24Intensive Care Unit, Alfred Hospital, Melbourne, Victoria, Australia. 25Department of Critical Care Medicine, Queen’s University and Kingston Health Sciences Centre, Kingston, Ontario, Canada. 26Department of Anaesthesia and Intensive Care, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong, China. 27Wellcome-Wolfson Institute for Experimental Medicine, Queen’s University Belfast, Belfast, UK. 28Department of Intensive Care Medicine, Royal Victoria Hospital, Belfast, UK. 29UCL Centre for Human Health and Performance, London, W1T 7HA, UK. 30Faculty of Medicine, University of São Paulo, São Paulo, Brazil. 31Medical Genetics, University of Siena, Siena, Italy. 32Genetica Medica, Azienda Ospedaliero-Universitaria Senese, Siena, Italy. 33Centre for Global Health Research, Usher Institute of Population Health Sciences and Informatics, Teviot Place, Edinburgh, EH8 9AG, UK. 34Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden. 35Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK. 36National Heart and Lung Institute, Imperial College London, London, UK. 37Imperial College Healthcare NHS Trust London, London, UK. 38Respiratory Medicine, Alder Hey Children’s Hospital, Institute in The Park, University of Liverpool, Alder Hey Children’s Hospital, Liverpool, UK. 325These authors contributed equally. Additionally, such studies invite the collaboration of researchers. *A list of authors and affiliations is available in the online version of the paper. ✉e-mail: j.k.baillie@ed.ac.uk
a. Gene-level Manhattan plot showing the raw p-value results from a cross-tissue meta-TWAS analysis (see Methods). The z-scores indicate the direction of effect of genotype-imputed expression of protein-coding gene transcripts in lung tissue (GTEX v8). The red highlights indicate genome-wide significance at p < 5 × 10^-6.
Table 1 Lead Variants from Independent Genome-Wide Significant Regions
| SNP | chr:pos(b37) | Risk | Alt | RAFgcc | RAFukb | OR | CI | Pgcc.ukb | Pgcc.gs | Pgcc.100k | Locus |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs73064425 | 3:45901089 | T | C | 0.15 | 0.07 | 2.1 | 1.88-2.45 | 4.8x10-30 | 2.9x10-27 | 3.6x10-32 | LZTFL1 |
| rs9380142 | 6:29798794 | A | G | 0.74 | 0.69 | 1.3 | 1.18-1.43 | 3.2x10-8 | 0.00091 | 1.8x10-8 | HLA-G |
| rs143334143 | 6:31121426 | A | G | 0.12 | 0.07 | 1.9 | 1.61-2.13 | 8.8x10-18 | 2.6x10-24 | 5.8x10-18 | CCHCR1 |
| rs3131294 | 6:32180146 | G | A | 0.9 | 0.86 | 1.5 | 1.28-1.66 | 2.8x10-8 | 1.3x10-10 | 2.3x10-8 | NOTCH4 |
| rs10735079 | 12:113380008 | A | G | 0.68 | 0.63 | 1.3 | 1.18-1.42 | 1.6x10-8 | 2.8x10-9 | 4.7x10-6 | OAS1/3 |
| rs2109069 | 19:4719443 | A | G | 0.38 | 0.32 | 1.4 | 1.25-1.48 | 4x10-12 | 4.5x10-7 | 2.4x10-8 | DPP9 |
| rs74956615 | 19:10427721 | A | T | 0.079 | 0.05 | 1.6 | 1.35-1.87 | 2.3x10-8 | 2.2x10-13 | 3.9x10-6 | TYK2 |
| rs2236757 | 21:34624917 | A | G | 0.34 | 0.28 | 1.3 | 1.17-1.41 | 5x10-8 | 8.9x10-5 | 8.3x10-7 | IFNAR2 |
chr:pos - Chromosome and position of the top SNP (build 37); Risk - Risk allele; Alt - Alternative allele; RAF - Risk allele frequency; OR - Effect size (odds ratio) of the risk allele in the GenOMICC EUR analysis; CI - 95% confidence interval of the odds ratio in the GenOMICC EUR cohort; P - p-value; Locus - Gene nearest to the top SNP. Subscript identifiers indicate the cohorts used for the cases: gcc - GenOMICC EUR; and the controls: ukb - UK Biobank; gs - Generation Scotland; 100k - 100,000 Genomes.
Table 2 Meta-analysis of overlapping SNPs between GenOMICC (EUR) and HGI (hospitalized Covid-19 vs. population) and 23andMe studies
| SNP | chr:pos(b37) | Risk | Alt | ORgcc | CIgcc | Pgcc | ORmeta | CImeta | Pmeta | Locus |
|---|---|---|---|---|---|---|---|---|---|---|
| rs71325088 | 3:45862952 | C | T | 2.1 | 1.87-2.43 | 9.3x10-30 | 1.9 | 1.73-2 | 2.5x10-54 | LZTFL1 |
| rs143334143 | 6:31121426 | A | G | 1.8 | 1.61-2.13 | 8.8x10-18 | 1.3 | 1.27-1.48 | 1.5x10-10 | CCHCR1 |
| rs6489867 | 12:113363550 | T | C | 1.3 | 1.15-1.37 | 6.9x10-7 | 1.2 | 1.14-1.25 | 9.7x10-10 | OAS1 |
| rs2109069 | 19:4719443 | A | G | 1.4 | 1.25-1.48 | 4x10-12 | 1.2 | 1.19-1.31 | 7x10-13 | DPP9 |
| rs11085727 | 19:10466123 | T | C | 1.3 | 1.17-1.4 | 1.3x10-7 | 1.2 | 1.18-1.31 | 1.2x10-13 | TYK2 |
| rs13050728 | 21:34615210 | T | C | 1.3 | 1.15-1.38 | 3x10-7 | 1.2 | 1.16-1.28 | 5.1x10-12 | IFNAR2 |
This is a meta-analysis of all available data; therefore, SNPs are included in this table if they meet a more stringent p-value threshold of p<10^-8, as an attempt at external replication cannot be made. SNP - the strongest SNP within the locus; Risk - risk allele; Alt - alternative allele; OR - odds ratio of the risk allele; CI - 95% confidence interval of the odds ratio; Locus - gene closest to the top SNP. Subscript identifiers denote gcc - GenOMICC study, European ancestry, comparison with UK Biobank, meta-analysis of European ancestry cases across all three studies (GenOMICC, HGI, 23andMe).
GenOMICC Consortium
GenOMICC Collaborators
Sara Clohisey1, Peter Horby21, Johnny Millar1, Julian Knight14, Hugh Montgomery29, David Maslove25, Lowell Ling26, Alistair Nichol22, Charlotte Summers15, Tim Walsh11, Charles Hinds20, Malcolm G. Semple18,38, Peter J.M. Openshaw36,37, Manu Shankar-Hari16, Antonia Ho19, Danny McAuley27,28, Chris Ponting2, Kathy Rowan7 & J. Kenneth Baillie1,2,11
Central management and laboratory team
Fiona Griffiths1, Wilna Oosthuyzen1, Jen Meikle1, Paul Finernan1, James Furniss1, Ellie Mcmaster1, Andy Law1, Sara Clohisey1, J. Kenneth Baillie1,11, Trevor Paterson1, Tony Wackett1, Ruth Armstrong1, Lee Murphy6, Angie Fawkes6, Richard Clark6, Audrey Coutts6, Lorna Donnelly6, Tammy Gilchrist6, Katarzyna Hafezi6, Louise Macgillivray6, Alan Maclean6, Sarah McCafferty6, Kirstie Morrice6, Jane Weaver1, Ceilia Boz1, Ailsa Golightly1, Mari Ward1, Hanning Mal1, Helen Szoor-McElhinney1, Adam Brown1, Ross Hendry1, Andrew Stenhouse1, Louise Cullum1, Dawn Law1, Sarah Law1, Rachel Law1, Max Head Fourman1, Maaike Swets1, Nicky Day1, Filip Taneski1, Esther Duncan1, Marie Zechner1 & Nicholas Parkinson1
Data analysis team
Erola Pairo-Castineira1,2, Sara Clohisey1, Lucija Klaric2, Andrew D. Bretherick2, Konrad Rawlik1, Dorota Pasko3, Susan Walker3, Nick Parkinson1, Max Head Fourman1, Clark D Russell1,4, James Furniss1, Anne Richmond2, Elvina Gountouna5, David Harrison7, Bo Wang1, Yang Wu8, Alison Meynert2, Athanasios Kousathanas3, Loukas Moutsianas3, Zhijian Yang9, Ranran Zhai9, Chenqing Zheng9, Graeme Grimes2, Jonathan Millar1, Barbara Shih1, Marie Zechner1, Jian Yang12,13, Xia Shen9,33,34, Chris P. Ponting2, Albert Tenesa1,2,33, Kathy Rowan7, Andrew Law1, Veronique Vitart2, James F. Wilson2,33, J. Kenneth Baillie1,2,11
Barts Health NHS Trust, London, UK
D. Collier39, S. Wood39, A. Zak39, C. Borra39, M. Matharu39, P. May39, Z. Alldis39, O. Mitchelmore39, R. Bowles39, A. Easthorpe39, F. Bibi39, I. Lancoma-Malcolm39, J. Gurasashvili39, J. Pheby39, J. Shiel39, M. Bolton39, M. Patel39, M. Taylor39, O. Zongo39, P. Ebano39, P. Harding39, R. Astin-Chamberlain39, Y. Choudhury39, A. Cox39, D. Kallon39, M. Burton39, R. Hall39, S. Blowes39, Z. Prime39, J. Biddle39, O. Prysyazhna39, T. Newman39, C. Tierney39 & J. Kassam39
Guys and St Thomas’ Hospital, London, UK
M. Shankar-Hari40, M. Ostermann40, S. Campos40, A. Bociek40, R. Lim40, N. Grau40, T. O. Jones40, C. Whitton40, M. Marotti40 & G. Arbane40
James Cook University Hospital, Middlesburgh, UK
S. Bonner41, K. Hugill41 & J. Reid41
The Royal Liverpool University Hospital, Liverpool, UK
I. Welters42, V. Waugh42, K. Williams42, D. Shaw42, J. Fernandez Roman42, M. Lopez Martinez42, E. Johnson42, A. Waite42, B. Johnson42, O. Hamilton42 & S. Mulla42
King’s College Hospital, London, UK
M. McPhail43 & J. Smith43
Royal Infirmary of Edinburgh, Edinburgh, UK
J. K. Baillie1,2,11, L. Barclay44, D. Hope44, C. McCulloch44, L. McQuillan44, S. Clark44, J. Singleton44, K. Priestley44, N. Rea44, M. Callaghan44, R. Campbell44, G. Andrew44 & L. Marshall44
John Radcliffe Hospital, Oxford, UK
S. McKechnie45, P. Hutton45, A. Bashyal45 & N. Davidson45
Addenbrooke’s Hospital, Cambridge, UK
C. Summers46, P. Polgarova46, K. Stroud46, N. Pathan46, K. Elston46 & S. Agrawal46
Morriston Hospital, Swansea, UK
C. Battle47, L. Newey47, T. Rees47, R. Harford47, E. Brinkworth47, M. Williams47 & C. Murphy47
Ashford and St Peter’s Hospital, Surrey, UK
I. White48 & M. Croft48
Royal Stoke University Hospital, Staffordshire, UK
N. Bandla49, M. Gellamucho49, J. Tomlinson49, H. Turner49, M. Davies49, A. Quinn49, I. Hussain49, C. Thompson49, H. Parker49, R. Bradley49 & R. Griffiths49
Queen Elizabeth Hospital, Birmingham, UK
J. Scriven50 & J. Gill50
Glasgow Royal Infirmary, Glasgow, UK
A. Puxty51, S. Cathcart51, D. Salutous51, L. Turner51, K. Duffy51 & K. Puxty51
Kingston Hospital, Surrey, UK
A. Joseph52, R. Herdman-Grant52, R. Simms52, A. Swain52, A. Naranjo52, R. Crowe52,K. Sollesta52, A. Loveridge52, D. Baptista52 & E. Morino52
The Tunbridge Wells Hospital and Maidstone Hospital, Kent, UK
M. Davey53, D. Golden53 & J. Jones53
North Middlesex University Hospital NHS Trust, London, UK
J. Moreno Cuesta54, A. Haldeos54, D. Bakthavatsalam54, R. Vincent54, M. Elhassan54,K. Xavier54, A. Ganesan54 & D. Purohit M. Abdelrazik54
Bradford Royal Infirmary, Bradford, UK
J. Morgan55, L. Akeroyd55, S. Bano55, D. Warren55, M. Bromley55, K. Sellick55, L. Gurr55,B. Wilkinson55, V. Nagarajan55 & P. Szedlak55
Blackpool Victoria Hospital, Blackpool, UK
J. Cupitt56, E. Stoddard56, L. Benham56, S. Preston56, N. Slawson56, Z. Bradshaw56, J. brown56,M. Caswell56 & S. Melling56
Countess of Chester Hospital, Chester, UK
P. Bamford57, M. Faulkner57, K. Cawley57, H. Jeffrey57, E. London57, H. Sainsbury57, I. Nagra57,F. Nasir57, Ce Dunmore57, R. Jones57, A. Abraheem57, M. Al-Moasseb57 & R. Girach57
Wythenshawe Hospital, Manchester, UK
C. Brantwood58, P. Alexander58, J. Bradley-Potts58, S. Allen58 & T. Felton58
St George’s Hospital, London, UK
S. Manna59, S. Farnell-Ward59, S. Leaver59, J. Queiroz59, E. Maccacari59, D. Dawson59, C. Castro Delgado59, R. Pepermans Saluzzio59, O. Ezeobu59, L. Ding59, C. Sicat59, R. Kanu59, G. Durrant59, J. Texeira59, A. Harrison59 & T. Samakomva59
Good Hope Hospital, Birmingham, UK
J. Scriven60, H. Willis60, B. Hopkins60 & L. Thrasyvoulou60
Stepping Hill Hospital, Stockport, UK
M. Jackson61, A. Zaki61, C. Tibke61, S. Bennett61, W. Woodyatt61, A. Kent61 & E. Goodwin61
Manchester Royal Infirmary, Manchester, UK
C. Brandwood62, R. Clark62 & L. Smith62
Royal Alexandra Hospital, Paisley, UK
K. Rooney63, N. Thomson63, N. Rodden63, E. Hughes63, D. McGlynn63, C. Clark63, P. Clark63, L. Abel63, R. Sundaram63, L. Gemmell63, M. Brett63, J. Hornsby63, P. MacGoey63, R. Price63, B. Digby63, P. O’Neil63, P. McConnell63 & P. Henderson63
Queen Elizabeth University Hospital, Glasgow, UK
S. Henderson64, M. Sim64, S. Kennedy-Hay64, C. McParland64, L. Rooney64 & N. Baxter64
Queen Alexandra Hospital, Portsmouth, UK
D. Pogson65, S. Rose65, Z. Daly65 & L. Brimfield65
BHRUT (Barking Havering) - Queens Hospital and King George Hospital, Essex, UK
M. K. Phull66, M. Hussain66, T. Pogreban66, L. Rosaroso66 & E. Salciute L. Grauslyte66
University College Hospital, London, UK
D. Brealey67, E. Wraith67, N. MacCallum67, G. Bercades67, I. Hass67, D. Smyth67, A. Reyes67 & G. Martir67
Royal Victoria Infirmary, Newcastle Upon Tyne, UK
I. D. Clement68, K. Webster68, C. Hays68 & A. Gulati68
Western Sussex Hospitals, West Sussex, UK
L. Hodgson69, M. Margarson69, R. Gomez69, Y. Baird69, Y. Thirlwall69, L. Folkes69, A. Butler69, E. Meadows69, S. Moore69, D. Raynard69, H. Fox69, L. Riddles69, K. King69, S. Kimber69, G. Hobden69, A. McCarthy69, V. Cannons69, I. Balagosa69, I. Chadbourn69 & A. Gardner69
Salford Royal Hospital, Manchester, UK
D. Horner70, D. McLaughlanv70, B. Charles70, N. Proudfoot70, T. Marsden70, L. Mc Morrow70, B. Blackledge70, J. Pendlebury70, A. Harvey70, E. Apetri70, C. Basikolo70, L. Catlow70, R. Doonan70, K. Knowles70, S. Lee70, D. Lomas70, C. Lyons70, J. Perez70, M. Poulaka70, M. Slaughter70, K. Slevin70, M. Taylor70, V. Thomas70, D. Walker70 & J. Harris70
The Royal Oldham Hospital, Manchester, UK
A. Drummond71, R. Tully71, J. Dearden71, J. Philbin71, S. Munt71, C. Rishton71, G. O’Connor71, M. Mulcahy71, E. Dobson71, J. Cuttler71 & M. Edward71
Pinderfields General Hospital, Wakefield, UK
A. Rose72, B. Sloan72, S. Buckley72, H. Brooke72, E. Smithson72, R. Charlesworth72, R. Sandu72, M. Thirumaran72, V. Wagstaff72 & J. Cebrian Suarez72
Basildon Hospital, Basildon, UK
A. Kaliappan73, M. Vertue73, A. Nicholson73, J. Riches73, A. Solesbury73, L. Kittridge73, M. Forsey73 & G. Maloney73
University Hospital of Wales, Cardiff, UK
J. Cole74, M. Davies74, R. Davies74, H. Hill74, E. Thomas74, A. Williams74, D. Duffin74 & B. Player74
Broomfield Hospital, Chelmsford, UK
J. Radhakrishnan75, S. Gibson75, A. Lyle75 & F. McNeela75
Royal Brompton Hospital, London, UK
B. Patel76, M. Gummadi76, G. Sloane76, N. Dormand76, S. Salmi76, Z. Farzad76, D. Cristiano76, K. Liyanage76, V. Thwaites76 & M. Varghese76
Nottingham University Hospital, Nottingham, UK
M. Meredith7
Royal Hallamshire Hospital and Northern General Hospital, Sheffield, UK
G. Mills78, J. Willson78, K. Harrington78, B. Lenagh78, K. Cawthron78, S. Masuko78, A. Raithatha78, K. Bauchmuller78, N. Ahmad78, J. Barker78, Y. Jackson78, F. Kibutu78 & S. Bird78
Royal Hampshire County Hospital, Hampshire, UK
G. Watson79, J. Martin79, E. Bevan79, C. Wrey Brown79 & D. Trodd79
Queens Hospital Burton, Burton-On-Trent, UK
K. English80, G. Bell80, L. Wilcox80 & A. Katary80
New Cross Hospital, Wolverhampton, UK
S. Gopal81, V. Lake81, N. Harris81, S. Metherell81 & E. Radford81
Heartlands Hospital, Birmingham, UK
J. Scriven82, F. Moore82, H. Bancroft82, J. Daglish82, M. Sangombe82, M. Carmody82, J. Rhodes82 & M. Bellamy82
Walsall Manor Hospital, Walsall, UK
A. Garg83, A. Kuravi83, E. Virgilio83, P. Ranga83, J. Butler83, L. Botfield83, C. Dexter83 & J. Fletcher83
Stoke Mandeville Hospital, Buckinghamshire, UK
P. Shanmugasundaram84, G. Hambrook84, I. Burn84, K. Manso84, D. Thornton84, J. Tebbutt84 & R. Penn84
Sandwell General Hospital, Birmingham, UK
J. Hulme85, S. Hussain85, Z. Maqsood85, S. Joseph85, J. Colley85, A. Hayes85, C. Ahmed85, R. Haque85, S. Clamp85, R. Kumar85, M. Purewal85 & B. Baines85
Royal Berkshire NHS Foundation Trust, Berkshire, UK
M. Frise86, N. Jacques86, H. Coles86, J. Caterson86, S. Gurung Rai86, M. Brunton86, E. Tilney86, L. Keating86 & A. Walden86
Charing Cross Hospital, St Mary’s Hospital and Hammersmith Hospital, London, UK
D. Antcliffe87, A. Gordon87, M. Templeton87, R. Rojo87, D. Banach87, S. Sousa Arias87, Z. Fernandez87 & P. Coghlan87
Dumfries and Galloway Royal Infirmary, Dumfries, UK
D. Williams88 & C. Jardine88
Bristol Royal Infirmary, Bristol, UK
J. Bewley89, K. Sweet89, L. Grimmer89, R. Johnson89, Z. Garland89 & B. Gumbrill89
Royal Sussex County Hospital, Brighton, UK
C. Phillips90, L. Ortiz-Ruiz de Gordoa90 & E. Peasgood90
Whiston Hospital, Prescot, UK
A. Tridente91 & K. Shuker S. Greer91
Royal Glamorgan Hospital, Cardiff, UK
C. Lynch92, C. Pothecary92, L. Roche92, B. Deacon92, K. Turner92, J. Singh92 & G. Sera Howe92
King’s Mill Hospital, Nottingham, UK
P. Paul93, M. Gill93, I. Wynter93, V. Ratnam93 & S. Shelton93
Fairfield General Hospital, Bury, UK
J. Naisbitt94 & J. Melville94
Western General Hospital, Edinburgh, UK
R. Baruah95 & S. Morrison95
Northwick Park Hospital, London, UK
A. McGregor96, V. Parris96, M. Mpelembue96, S. Srikaran96, C. Dennis96 & A. Sukha96
Royal Preston Hospital, Preston, UK
A. Williams97 & M. Verlande97
Royal Derby Hospital, Derby, UK
K. Holding98, K. Riches98, C. Downes98 & C. Swan98
Sunderland Royal Hospital, Sunderland, UK
A. Rostron99, A. Roy99, L. Woods99, S. Cornell99 & F. Wakinshaw99
Royal Surrey County Hospital, Guildford, UK
B. Creagh-Brown100, H. Blackman100, A. Salberg100, E. Smith100, S. Donlon100, S. Mtuwa100,
N. Michalak-Glinska100, S. Stone100, C. Beazley100 & V. Pristopan100
Derriford Hospital, Plymouth, UK
N. Nikitas101, L. Lankester101 & C. Wells101
Croydon University Hospital, Croydon, UK
A. S. Raj102, K. Fletcher102, R. Khade102 & G. Tsinaslanidis102
Victoria Hospital, Kirkcaldy, UK
M. McMahon103, S. Fowler103, A. McGregor103 & T. Coventry103
Milton Keynes University Hospital, Milton Keynes, UK
R. Stewart104, L. Wren104, E. Mwaura104, L. Mew104, A. Rose104, D. Scaletta104 & F. Williams104
Barnsley Hospital, Barnsley, UK
K. Inweregbu105, A. Nicholson105, N. Lancaster105, M. Cunningham105, A. Daniels105, L. Harrison105, S. Hope105, S. Jones105, A. Crew105, G. Wray105, J. Matthews105 & R. Crawley105
York Hospital, York, UK
J. Carter106, I. Birkinshaw106, J. Ingham106, Z. Scott106, K. Howard106, R. Joy106 & S. Roche106
University Hospital of North Tees, Stockton on Tees, UK
M. Clark107 & S. Purvis107
University Hospital Wishaw, Wishaw, UK
A. Morrison108, D. Strachan108, M. Taylor108, S. Clements108 & K. Black108
Whittington Hospital, London, UK
C. Parmar109, A. Altabaibeh109, K. Simpson109, L. Mostoles109, K. Gilbert109, L. Ma109 & A. Alvaro109
Southmead Hospital, Bristol, UK
M. Thomas110, B. Faulkner110, R. Worner110, K. Hayes110, E. Gendall110, H. Blakemore110, B. Borislavova110 & E. Goff110
The Royal Papworth Hospital, Cambridge, UK
A. Vuylsteke111, L. Mwaura111, J. Zamikula111, L. Garner111, A. Mitchell111, S. Mepham111, L. Cagova111, A. Fofano111, H. Holcombe111 & K. Praman111
Royal Gwent Hospital, Newport, UK
T. Szakmany112, A. E. Heron112, S. Cherian112, S. Cutler112 & A. Roynon-Reed112
Norfolk and Norwich University hospital (NNUH), Norwich, UK
G. Randell113, K. Convery113, K. Stammers D. Fottrell-Gould113, L. Hudig113 & J. Keshet-price113
Great Ormond St Hospital and UCL Great Ormond St Institute of Child Health NIHR
Biomedical Research Centre, London, UK
M. Peters114, L. O’Neill114, S. Ray114, H. Belfield114, T. McHugh114, G. Jones114, O. Akinkugbe114, A. Tomas114, E. Abaleke114, E. Beech114, H. Meghari114, S. Yussuf114 & A. Bamford114
Airedale General Hospital, Keighley, UK
B. Hairsine115, E. Dooks115, F. Farquhar115, S. Packham115, H. Bates115, C. McParland115 & L. Armstrong115
Aberdeen Royal Infirmary, Aberdeen, UK
C. Kaye116, A. Allan116, J. Medhora116, J. Liew116, A. Botello116 & F. Anderson116
Southampton General Hospital, Southampton, UK
R. Cusack117, H. Golding117, K. Prager117, T. Williams117, S. Leggett117, K. Golder117, M. Male117, O. Jones117, K. Criste117 & M. Marani117
Russell’s Hall Hospital, Dudley, UK
Dr Anumakonda118, V. Amin118, K. Karthik118, R. Kausar118, E. Anastasescu118, K. Reid118 & Ms Jacqui118
Rotherham General Hospital, Rotherham, UK
A. Hormis119, R. Walker119 & D. Collier119
North Manchester General Hospital, Manchester, UK
T. Duncan120, A. Uriel120, A. Ustianowski120, H. T-Michael120, M. Bruce120, K. Connolly120 & K. Smith120
Basingstoke and North Hampshire Hospital, Basingstoke, UK
R. Partridge121, D. Griffin121, M. McDonald121 & N. Muchenje121
Royal Free Hospital, London, UK
D. Martin122, H. Filipe122, C. Eastgate122 & C. Jackson122
Hull Royal Infirmary, Hull, UK
A. Gratrix123, L. Foster123, V. Martinson123, E. Stones123, Caroline Abernathy123 & P. Parkinson123
Harefield Hospital, London, UK
A. Reed124, C. Prendergast124, P. Rogers124, M. Woodruff124, R. Shokkar124, S. Kaul124, A. Barron124 & C. Collins124
Chesterfield Royal Hospital Foundation Trust, Chesterfield, UK
S. Beavis125, A. Whileman125, K. Dale125, J. Hawes125, K. Pritchard125, R. Gascoyne125 & L. Stevenson125
Barnet Hospital, London, UK
R. Jha126, L. Lim126 & V. Krishnamurthy126
Aintree University Hospital, Liverpool, UK
R. Parker127, I. Turner-Bone127, L. Wilding127 & A. Reddy127
St James’s University Hospital and Leeds General Infirmary, Leeds, UK
S. Whiteley128, E. Wilby128, C. Howcroft128, A. Aspinwall128, S. Charlton128 & B. Ogg128
Glan Clwyd Hospital, Bodelwyddan, UK
D. Menzies129, R. Pugh129, E. Allan129, R. Lean129, F. Davies129, J. Easton129, X. Qiu129, S. Kumar129 & K. Darlington129
University Hospital Crosshouse, Kilmarnock, UK
G. Houston130, P. O’Brien130, T. Geary130, J. Allan130 & A. Meikle130
Royal Bolton Hospital, Bolton, UK
G. Hughes131, M. Balasubramaniam131, S. Latham131, E. McKenna131 & R. Flanagan131
Princess of Wales Hospital, Llantrisant, UK
S. Sathe132, E. Davies132 & L. Roche132
Pilgrim Hospital, Lincoln, UK
M. Chablani133, A. Kirkby133, K. Netherton133 & S. Archer133
Northumbria Healthcare NHS Foundation Trust, North Shields, UK
B. Yates134 & C. Ashbrook-Raby134
Ninewells Hospital, Dundee, UK
S. Cole135, M. Casey135, L. Cabrelli135, S. Chapman135, M. Casey135, A. Hutcheon135, C. Whyte135 & C. Almaden-Boyle135
Lister Hospital, Stevenage, UK
N. Pattison136 & C. Cruz136
Bedford Hospital, Bedford, UK
A. Vochin137, H. Kent137, A. Thomas137, S. Murdoch137, B. David137, M. Penacerrada137, G. Lubimbi137, V. Bastion137, R. Wulandari137, J. Valentine137 & D. Clarke137
Royal United Hospital, Bath, UK
A. Serrano-Ruiz138, S. Hierons138, L. Ramos138, C. Demetriou138, S. Mitchard138 & K. White138
Royal Bournemouth Hospital, Bournemouth, UK
N. White139, S. Pitts139, D. Branney139 & J. Frankham139
The Great Western Hospital, Swindon, UK
M. Watters140, H. Langton140 & R. Prout140
Watford General Hospital, Watford, UK
V. Page141 & T. Varghes141
University Hospital North Durham, Darlington, UK
A. Cowton142, A. Kay142, K. Potts142, M. Birt142, M. Kent142 & A. Wilkinson142
Tameside General Hospital, Ashton Under Lyne, UK
E. Jude143, V. Turner143, H. Savill143, J. McCormick143, M. Clark143, M. Coulding143, S. Siddiqui143, O. Mercer143, H. Rehman143 & D. Potla143
Princess Royal Hospital, Telford and Royal Shrewsbury Hospital, Shrewsbury, UK
N. Capps144, D. Donaldson144, J. Jones144, H. Button144, T. Martin144, K. Hard144, A. Agasou144, L. Tonks144, T. Arden144, P. Boyle144, M. Carnahan144, J. Strickley144, C. Adams144, D. Childs144, R. Rikunenko144, M. Leigh144, M. Breekes144, R. Wilcox144, A. Bowes144, H. Tiveran144,
F. Hurford144, J. Summers144, A. Carter144, Y. Hussain144, L. Ting144, A. Javaid144, N. Motherwell144, H. Moore144, H. Millward144, S. Jose144, N. Schunki144, A. Noakes144 & C. Clulow144
Arrowe Park Hospital, Wirral, UK
G. Sadera145, R. Jacob145 & C. Jones145
The Queen Elizabeth Hospital, King’s Lynn, UK
M. Blunt146, Z. Coton146, H. Curgenven146, S. Mohamed Ally146, K. Beaumont146, M. Elsaadany146, K. Fernandes146, I. Ali Mohamed Ali146, H. Rangarajan146, V. Sarathy146, S. Selvanayagam146, D. Vedage146 & M. White146
Royal Blackburn Teaching Hospital, Blackburn, UK
M. Smith147, N. Truman147, S. Chukkambotla147, S. Keith147, J. Cockerill-Taylor147, J. Ryan-Smith147, R. Bolton147, P. Springle147, J. Dykes147, J. Thomas147, M. Khan147, M. T. Hijazi147, E. Massey147 & G. Croston147
Poole Hospital, Poole, UK
H. Reschreite r148, J. Camsooksai148, S. Patch148, S. Jenkins148, C. Humphrey148, B. Wadams148 & J. Camsooksai148
Medway Maritime Hospital, Gillingham, UK
N. Bhatia149, M. Msiska149 & O. Adanini149
Warwick Hospital, Warwick, UK
B. Attwood150 & P. Parsons150
The Royal Marsden Hospital, London, UK
K. Tatham151, S. Jhanji151, E. Black151, A. Dela Rosa151, R. Howle151, B. Thomas151, T. Bemand151 & R. Raobaikady151
The Princess Alexandra Hospital, Harlow, UK
R. Saha152, N. Staines152, A. Daniel152 & J. Finn152
Musgrove Park Hospital, Taunton, UK
J. Hutter153, P. Doble153, C. Shovelton153 & C. Pawley153
George Eliot Hospital NHS Trust, Nuneaton, UK
T. Kannan154 & M. Hill154
East Surrey Hospital, Redhill, UK
E. Combes155, S. Monnery155 & T. Joefield155
West Middlesex Hospital, Isleworth, UK
M. Popescu156, M. Thankachen156 & M. Oblak156
Warrington General Hospital, Warrington, UK
J. Little157, S. McIvor157, A. Brady157, H. Whittle157, H. Prady157 & R. Chan157
Southport and Formby District General Hospital, Ormskirk, UK
A. Ahmed158 & A. Morris158
Royal Devon and Exeter Hospital, Exeter, UK
C. Gibson159, E. Gordon159, S. Keenan159, H. Quinn159, S. Benyon159, S. Marriott159, L. Zitter159, L. Park159 & K. Baines159
Macclesfield District General Hospital, Macclesfield, UK
M. Lyons160, M. Holland160, N. Keenan160 & M. Young160
Borders General Hospital, Melrose, UK
S. Garrioch161, J. Dawson161 & M. Tolson161
Birmingham Children’s Hospital, Birmingham, UK
B. Scholefield162 & R. Bi162
William Harvey Hospital, Ashford, UK
N. Richardson163, N. Schumacher163, T. Cosier163 & G. Millen163
Royal Lancaster Infirmary, Lancaster, UK
A. Higham164 & K. Simpson164
Queen Elizabeth the Queen Mother Hospital, Margate, UK
S. Turki165, L. Allen165, N. Crisp165, T. Hazleton165, A. Knight165, J. Deery165, C. Price165, S. Turney165, S. Tilbey165 & E. Beranova165
Liverpool Heart and Chest Hospital, Liverpool, UK
D. Wright166, L. Georg166 & S. Twiss166
Darlington Memorial Hospital, Darlington, UK
A. Cowton167, S. Wadd167 & K. Postlethwaite167
Southend University Hospital, Westcliff-on-Sea, UK
P. Gondo168, B. Masunda168, A. Kayani168 & B. Hadebe168
Raigmore Hospital, Inverness, UK
J. Whiteside169, R. Campbell169 & N. Clarke169
Salisbury District Hospital, Salisbury, UK
P. Donnison170, F. Trim170 & I. Leadbitter170
Peterborough City Hospital, Peterborough, UK
D. Butcher171 & S. O’Sullivan171
Ipswich Hospital, Ipswich, UK
B. Purewal172, B. Purewal172, S. Bell172 & V. Rivers172
Hereford County Hospital, Hereford, UK
R. O’Leary173, J. Birch173, E. Collins173, S. Anderson173, K. Hammerton173 & E. Andrews173
Furness General Hospital, Barrow-in-Furness, UK
A. Higham174 & K. Burns174
Forth Valley Royal Hospital, Falkirk, UK
I. Edmond175, D. Salutous175, A. Todd175, J. Donnachie175, P. Turner175, L. Prentice175, L. Symon175, N. Runciman175 & F. Auld175
Torbay Hospital, Torquay, UK
M. Halkes176, P. Mercer176 & L. Thornton176
St Mary’s Hospital, Newport, UK
G. Debreceni177, J. Wilkins177, A. Brown177 & V. Crickmore177
Royal Manchester Children’s Hospital, Manchester, UK
G. Subramanian178, R. Marshall178, C. Jennings178, M. Latif178 & L. Bunni178
Royal Cornwall Hospital, Truro, UK
M. Spivey179, S. Bean179 & K. Burt179
Queen Elizabeth Hospital Gateshead, Gateshead, UK
V. Linnett180, J. Ritzema180, A. Sanderson180, W. McCormick180 & M. Bokhari180
Kent & Canterbury Hospital, Canterbury, UK
R. Kapoor181 & D. Loader181
James Paget University Hospital NHS Trust, Great Yarmouth, UK
A. Ayers182, W. Harrison182 & J. North182
Darent Valley Hospital, Dartford, UK
Z. Belagodu183, R. Parasomthy183, O. Olufuwa183, A. Gherman183, B. Fuller183 & C. Stuart183
The Alexandra Hospital, Redditch and Worcester Royal Hospital, Worcester, UK
O. Kelsall184, C. Davis184, L. Wild184, H. Wood184, J. Thrush184, A. Durie184, K. Austin184, K. Archer184, P. Anderson184 & C. Vigurs184
Ysbyty Gwynedd, Bangor, UK
C. Thorpe185, A. Thomas185, E. Knights185, N. Boyle185 & A. Price185
Yeovil Hospital, Yeovil, UK
A. Kubisz-Pudelko186, D. Wood186, A. Lewis186, S. Board186, L. Pippard186, J. Perry186 & K. Beesley186
University Hospital Hairmyres, East Kilbride, UK
A. Rattray187, M. Taylor187, E. Lee187, L. Lennon187, K. Douglas187, D. Bell187, R. Boyle187 & L. Glass187
Scunthorpe General Hospital, Scunthorpe, UK
M. Nauman Akhtar188, K. Dent188, D. Potoczna188, S. Pearson188, E. Horsley188 & S. Spencer188
Princess Royal Hospital Brighton, West Sussex, UK
C. Phillips189, D. Mullan189, D. Skinner189, J. Gaylard189 & L. Ortiz-Ruizdegordoa189
Lincoln County Hospital, Lincoln, UK
R. Barber190, C. Hewitt190, A. Hilldrith190, S. Shepardson190, M. Wills190 &
K. Jackson-Lawrence190
Homerton University Hospital, London, UK
A. Gupta191, A. Easthope191, E. Timlick191 & C. Gorman191
Glangwili General Hospital, Camarthen, UK
I. Otaha192, A. Gales192, S. Coetzee192, M. Raj192 & M. Peiu192
Ealing Hospital, Southall, UK
V. Parris193, S. Quaid193 & E. Watson193
Scarborough General Hospital, Scarborough, UK
K. Elliott194, J. Mallinson194, B. Chandler194 & A. Turnbull194
Royal Albert Edward Infirmary, Wigan, UK
A. Quinn195, C. Finch195, C. Holl195, J. Cooper195 & A. Evans195
Queen Elizabeth Hospital, Woolwich, London, UK
W. Khaliq196, A. Collins196 & E. Treus Gude196
North Devon District Hospital, Barnstaple, UK
N. Love197, L. van Koutrik197, J. Hunt197, D. Kaye197, E. Fisher197, A. Brayne197, V. Tuckey197, P. Jackson197 & J. Parkin197
National Hospital for Neurology and Neurosurgery, London, UK
D. Brealey198, E. Raith198, A. Tariq198, H. Houlden198, A. Tucci198, J. Hardy198 & E. Moncur198
Eastbourne District General Hospital, East Sussex, UK and Conquest Hospital, East Sussex, UK
J. Highgate199 & A. Cowley199
Diana Princess of Wales Hospital, Grimsby, UK
A. Mitra200, R. Stead200, T. Behan200, C. Burnett200, M. Newton200, E. Heeney200, R. Pollard200 & J. Hatton200
The Christie NHS Foundation Trust, Manchester, UK
A. Patel201, V. Kasipandian201, S. Allibone201 & R. M. Genetu201
Prince Philip Hospital, Lianelli, UK
I. Otahal202, L. O’Brien202, Z. Omar202, E. Perkins202 & K. Davies202
Prince Charles Hospital, Merthyr Tydfil, UK
D. Tetla203, C. Pothecary203 & B. Deacon203
Golden Jubilee National Hospital, Clydebank, UK
B. Shelley204 & V. Irvine204
Dorset County Hospital, Dorchester, UK
S. Williams205, P. Williams205, J. Birch205, J. Goodsell205, R. Tutton205, L. Bough205 & B. Winter-Goodwin205
Calderdale Royal Hospital, Halifax, UK
R. Kitson206, J. Pinnell206, A. Wilson206, T. Nortcliffe206, T. Wood206, M. Home206, K. Holdroyd206, M. Robinson206, K. Hanson206, R. Shaw206, J. Greig206, M. Brady206, A. Haigh206, L. Matupe206, M. Usher206, S. Mellor206, S. Dale206, L. Gledhill206, L. Shaw206, G. Turner206, D. Kelly206, B. Anwar206, H. Riley206, H. Sturgeon206, A. Ali206, L. Thomis206, D. Melia206,
A. Dance206 & K. Hanson206
West Suffolk Hospital, Suffolk, UK
S. Humphreys207, I. Frost207, V. Gopal207, J. Godden207, A. Holden207 & S. Swann207
West Cumberland Hospital, Whitehaven, UK
T. Smith208, M. Clapham208, U. Poultney208, R. Harper208 & P. Rice208
University Hospital Lewisham, London, UK
W. Khaliq209, R. Reece-Anthony209 & B. Gurung209
St John’s Hospital Livingston, Livingston, UK
S. Moultrie210 & M. Odam210
Sheffield Children’s Hospital, Sheffield, UK
A. Mayer211, A. Bellini211, A. Pickard211, J. Bryant211, N. Roe211 & J. Sowter211
Hinchingbrooke Hospital, Huntingdon, UK
D. Butcher212, K. Lang212 & J. Taylor212
Glenfield Hospital, Leicester, UK
P. Barry213
Bronglais General Hospital, Aberystwyth, UK
M. Hobrok214, H. Tench214, R. Wolf-Roberts214, H. McGuinness214 & R. Loosley214
Alder Hey Children’s Hospital, Liverpool, UK
D. Hawcutt215, L. Rad215, L. O’Malley215, P. Saunderson215, G. Seddon215, T. Anderson215 & N. Rogers215
University Hospital Monklands, Airdrie, UK
J. Ruddy216, H. Margaret216, M. Taylor216, C. Beith216, A. McAlpine216, L. Ferguson216, P. Grant216, S. MacFadyen216, M. McLaughlin216, T. Baird216, S. Rundell216, L. Glass216, B. Welsh216, R. Hamill216 & F. Fisher216
Cumberland Infirmary, Carlisle, UK
T. Smith217, J. Gregory217 & A. Brown217
39Barts Health NHS Trust, London, UK. 40Guys and St Thomas’ Hospital, London, UK. 41James Cook University Hospital, Middlesburgh, UK. 42The Royal Liverpool University Hospital, Liverpool, UK. 43King’s College Hospital, London, UK. 44Royal Infirmary of Edinburgh, Edinburgh, UK. 45John Radcliffe Hospital, Oxford, UK. 46Addenbrooke’s Hospital, Cambridge, UK. 47Morriston Hospital, Swansea, UK. 48Ashford and St Peter’s Hospital, Chertsey, UK. 49Royal Stoke University Hospital, Stoke-on-Trent, UK. 50Queen Elizabeth Hospital, Birmingham, UK. 51Glasgow Royal Infirmary, Glasgow, UK. 52Kingston Hospital, Kingston upon Thames, UK. 53The Tunbridge Wells Hospital and Maidstone Hospital, Maidstone, UK. 54North Middlesex University Hospital NHS Trust, London, UK. 55Bradford Royal Infirmary, Bradford, UK. 56Blackpool Victoria Hospital, Blackpool, UK. 57Countess of Chester Hospital, Chester, UK. 58Wythenshawe Hospital, Manchester, UK. 59St George’s Hospital, London, UK. 60Good Hope Hospital, Birmingham, UK. 61Stepping Hill Hospital, Stockport, UK. 62Manchester Royal Infirmary, Manchester, UK. 63Royal Alexandra Hospital, Paisley, UK. 64Queen Elizabeth University Hospital, Glasgow, UK. 65Queen Alexandra Hospital, Portsmouth, UK. 66BHRUT (Barking Havering) - Queens Hospital and King George Hospital, Ilford, UK. 67University College Hospital, London, UK. 68Royal Victoria Infirmary, Newcastle Upon Tyne, UK. 69Western Sussex Hospitals, Chichester, UK. 70Salford Royal Hospital, Manchester, UK. 71The Royal Oldham Hospital, Manchester, UK. 72Pinderfields General Hospital, Wakefield, UK. 73Basildon Hospital, Basildon, UK. 74University Hospital of Wales, Cardiff, UK. 75Broomfield Hospital, Chelmsford, UK. 76Royal Brompton Hospital, London, UK. 77Nottingham University Hosp tal, Nottingham, UK. 78Royal Hallamshire Hospital and Northern General Hospital, Sheffield, UK. 79Royal Hampshire County Hospital, Winchester, UK. 80Queens Hospital Burton, Burton-On-Trent, UK. 81New Cross Hospital, Wolverhampton, UK. 82Heartlands Hospital, Birmingham, UK. 83Walsall Manor Hospital, Walsall, UK. 84Stoke Mandeville Hospital, Aylesbury, UK. 85Sandwell General Hospital, Birmingham, UK. 86Royal Berkshire NHS Foundation Trust, Reading, UK. 87Charing Cross Hospital, St Mary’s Hospital and Hammersmith Hospital, London, UK. 88Dumfries and Galloway Royal Infirmary, Dumfries, UK. 89Bristol Royal Infirmary, Bristol, UK. 90Royal Sussex County Hospital, Brighton, UK. 91Whiston Hospital, Prescot, UK. 92Royal Glamorgan Hospital, Cardiff, UK. 93King’s Mill Hospital, Nottingham, UK. 94Fairfield General Hospital, Bury, UK. 95Western General Hospital, Edinburgh, UK. 96Northwick Park Hospital, London, UK. 97Royal Preston Hospital, Preston, UK. 98Royal Derby Hospital, Derby, UK. 99Sunderland Royal Hospital, Sunderland, UK. 100Royal Surrey County Hospital, Guildford, UK. 101Derriford Hospital, Plymouth, UK. 102Croydon University Hospital, Croydon, UK. 103Victoria Hospital, Kirkcaldy, UK. 104Milton Keynes University Hospital, Milton Keynes, UK. 105Barnsley Hospital, Barnsley, UK. 106York Hospital, York, UK. 107University Hospital of North Tees, Stockton on Tees, UK. 108University Hospital Wishaw, Wishaw, UK. 109Whittington Hospital, London, UK. 110Southmead Hospital, Bristol, UK. 111The Royal Papworth Hospital, Cambridge, UK. 112Royal Gwent Hospital, Newport, UK. 113Norfolk and Norwich University Hospital (NNUH), Norwich, UK. 114Great Ormond St Hospital and UCL Great Ormond St Institute of Child Health NIHR Biomedical Research Centre, London, UK. 115Airedale General Hospital, Keighley, UK. 116Aberdeen Royal Infirmary, Aberdeen, UK. 117Southampton General Hospital, Southampton, UK. 118Russell’s Hall Hospital, Dudley, UK. 119Rotherham General Hospital, Rotherham, UK. 120North Manchester General Hospital, Manchester, UK. 121Basingstoke and North Hampshire Hospital, Basingstoke, UK. 122Royal Free Hospital, London, UK. 123Hull Royal Infirmary, Hull, UK. 124Harefield Hospital, London, UK. 125Chesterfield Royal Hospital Foundation Trust, Chesterfield, UK. 126Barnet Hospital, London, UK. 127Aintree University Hospital, Liverpool, UK. 128St James’s University Hospital and Leeds General Infirmary, Leeds, UK. 129Glan Clwyd Hospital, Bodelwyddan, UK. 130University Hospital Crosshouse, Kilmarnock, UK. 131Royal Bolton Hospital, Bolton, UK. 132Princess of Wales Hospital, Llantrisant, UK. 133Pilgrim Hospital, Lincoln, UK. 134Northumbria Healthcare NHS Foundation Trust, North Shields, UK. 135Ninewells Hospital, Dundee, UK. 136Lister Hospital, Stevenage, UK. 137Bedford Hospital, Bedford, UK. 138Royal United Hospital, Bath, UK. 139Royal Bournemouth Hospital, Bournemouth, UK. 140The Great Western Hospital, Swindon, UK. 141Watford General Hospital, Watford, UK. 142University Hospital North Durham, Darlington, UK. 143Tameside General Hospital, Ashton Under Lyne, UK. 144Princess Royal Hospital Shrewsbury and Royal Shrewsbury Hospital, Shrewsbury, UK. 145Arrowe Park Hospital, Wirral, UK. 146The Queen Elizabeth Hospital, King’s Lynn, UK. 147Royal Blackburn Teaching Hospital, Blackburn, UK. 148Poole Hospital, Poole, UK. 149Medway Maritime Hospital, Gillingham, UK. 150Warwick Hospital, Warwick, UK. 151The Royal Marsden Hospital, London, UK. 152The Princess Alexandra Hospital, Harlow, UK. 153Musgrove Park Hospital, Taunton, UK. 154George Eliot Hospital NHS Trust, Nuneaton, UK. 155East Surrey Hospital, Redhill, UK. 156West Middlesex Hospital, Isleworth, UK. 157Warrington General Hospital, Warrington, UK. 158Southport and Formby District General Hospital, Ormskirk, UK. 159Royal Devon and Exeter Hospital, Exeter, UK. 160Macclesfield District General Hospital, Macclesfield, UK. 161Borders General Hospital, Melrose, UK. 162Birmingham Children’s Hospital, Birmingham, UK. 163William Harvey Hospital, Ashford, UK. 164Royal Lancaster Infirmary, Lancaster, UK. 165Queen Elizabeth the Queen Mother Hospital, Margate, UK. 166Liverpool Heart and Chest Hospital, Liverpool, UK. 167Darlington Memorial Hospital, Darlington, UK. 168Southend University Hospital, Westcliffon- Sea, UK. 169Raigmore Hospital, Inverness, UK. 170Salisbury District Hospital, Salisbury, UK. 171Peterborough City Hospital, Peterborough, UK. 172Ipswich Hospital, Ipswich, UK. 173Hereford County Hospital, Worcester, UK. 174Furness General Hospital, Barrow-in-Furness, UK. 175Forth Valley Royal Hospital, Falkirk, UK. 176Torbay Hospital, Torquay, UK. 177St Mary’s Hospital, Newport, UK. 178Royal Manchester Children’s Hospital, Manchester, UK. 179Royal Cornwall Hospital, Truro, UK. 180Queen Elizabeth Hospital Gateshead, Gateshead, UK. 181Kent & Canterbury Hospital, Canterbury, UK. 182James Paget University Hospital NHS Trust, Great Yarmouth, UK. 183Darent Valley Hospital, Dartford, UK. 184The Alexandra Hospital, Redditch and Worcester Royal Hospital, Worcester, UK. 185Ysbyty Gwynedd, Bangor, UK. 186Yeovil Hospital, Yeovil, UK. 187University Hospital Hairmyres, East Kilbride, UK. 188Scunthorpe General Hospital, Scunthorpe, UK. 189Princess Royal Hospital Brighton, Haywards Heath, UK. 190Lincoln County Hospital, Lincoln, UK. 191Homerton University Hospital, London, UK. 192Glangwili General Hospital, Camarthen, UK. 193Ealing Hospital, London, UK. 194Scarborough General Hospital, Scarborough, UK. 195Royal Albert Edward Infirmary, Wigan, UK. 196Queen Elizabeth Hospital, Woolwich, London, UK. 197North Devon District Hospital, Barnstaple, UK. 198National Hospital for Neurology a d Neurosurgery, London, UK. 199Eastbourne District General Hospital, East Sussex, UK and Conquest Hospital, Eastbourne, UK. 200Diana Princess of Wales Hospital, Grimsby, UK. 201The Christie NHS Foundation Trust, Manchester, UK. 202Prince Philip Hospital, Lianelli, UK. 203Prince Charles Hospital, Merthyr Tydfil, UK. 204Golden Jubilee National Hospital, Clydebank, UK. 205Dorset County Hospital, Dorchester, UK. 206Calderdale Royal Hospital, Halifax, UK. 207West Suffolk Hospital, Bury St Edmunds, UK. 208West Cumberland Hospital, Whitehaven, UK. 209University Hospital Lewisham, London, UK. 210St John’s Hospital Livingston, Livingston, UK. 211Sheffield Children’s Hospital, Sheffield, UK. 212Hinchingbrooke Hospital, Huntingdon, UK. 213Glenfield Hospital, Leicester, UK. 214Bronglais General Hospital, Aberystwyth, UK. 215Alder Hey Children’s Hospital, Liverpool, UK. 216University Hospital Monklands, Airdrie, UK. 217Cumberland Infirmary, Carlisle, UK.
The list of members and their affiliations can be found in the supplementary information.
ISARIC4C Consortium
ISARICC Collaborators
J. Kenneth Baillie1,2,11, Malcolm G. Semple18,38, Peter J. M. Openshaw36,37, Gail Carson286, Beatrice Alex287, Benjamin Bach287, Wendy S. Barclay288, Debby Bogaert4, Meera Chand289, Graham S. Cooke290, Annemarie B. Docherty11,291, Jake Dunning36,292, Ana da Silva Filipe293, Tom Fletcher294, Christoper A. Green295, Ewen M. Harrison291, Julian A. Hiscox296, Antonia Ying Wai Ho293,297, Peter W. Horby21, Samreen Ijaz298, Saye Khoo299, Paul Klenerman300,301, Andrew Law1, Wei Shen Lim77, Alexander J. Mentzer300,302, Laura Merson286, Alison M. Meynert2, Mahdad Noursadeghi303, Shona C. Moore304, Massimo Palmarini293, William A. Paxton304,305, Georgios Pollakis304,305, Nicholas Price306,307, Andrew Rambaut308, David L. Robertson293, Clark D. Russell1,4, Vanessa Sancho-Shimizu309, Janet T. Scott293,310, Thushan de Silva311, Louise Sigfrid286, Tom Solomon18,312, Shiranee Sriskandan290,313, David Stuart314, Charlotte Summers15, Richard S. Tedder315,316,317, Emma C. Thomson293, AA Roger Thompson318, Ryan S. Thwaites36, Lance CW Turtle18,319 & Maria Zambon292 Project management team
Hayley Hardwick18, Chloe Donohue320, Ruth Lyons1, Fiona Griffiths1 & Wilna Oosthuyzen1
Data Analysis Team
Lisa Norman291, Riinu Pius291, Thomas M. Drake291, Cameron J. Fairfield291, Stephen R. Knight291, Kenneth A. Mclean291, Derek Murphy291 & Catherine A. Shaw291
Data Architecture Team
Jo Dalton320, Michelle Girvan320, Egle Saviciute320, Stephanie Roberts320, Janet Harrison320, Laura Marsh320, Marie Connor320, Sophie Halpin320, Clare Jackson320, Carrol Gamble320, Gary Leeming321, Andrew Law1, Murray Wham2, Sara Clohisey1, Ross Hendry1 & James Scott-Brown287
Data Analytics and Management Team
William Greenhalf322, Victoria Shaw323, Sara McDonald293 & Seán Keating11
286ISARIC Global Support Centre, Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK. 287School of Informatics, University of Edinburgh, Edinburgh, UK. 288Section of Molecular Virology, Imperial College London, London, UK. 289Antimicrobial Resistance and Hospital Acquired Infection Department, Public Health England, London, UK. 290Department of Infectious Disease, Imperial College London, London, UK. 291Centre for Medical Informatics, The Usher Institute, University of Edinburgh, Edinburgh, UK. 292National Infection Service, Public Health England, London, UK. 293MRC-University of Glasgow Centre for Virus Research, 464 Bearsden Road, Glasgow, UK. 294Liverpool School of Tropical Medicine, Liverpool, UK. 295Institute of M crobiology and Infection, University of Birmingham, Birmingham, UK. 296Institute of Infection and Global Health, University of Liverpool, Liverpool, UK. 297Department of Infectious Diseases, Queen Elizabeth University Hospital, Glasgow, UK. 298Virology Reference Department, National Infection Service, Public Health England, Colindale Avenue, London, UK. 299Department of Pharmacology, University of Liverpool, Liverpool, UK. 300Nuffield Department of Medicine, John Radcliffe Hospital, Oxford, UK. 301Translational Gastroenterology Unit, Nuffield Department of Medicine, University of Oxford, UK. 302Department of Microbiology/Infectious Diseases, Oxford University Hospitals NHS Foundation Trust, John Radcliffe Hospital, Oxford, UK. 303Division of Infection and Immunity, University College London, UK. 304Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, UK. 305NIHR Health Protection Research Unit in Emerging and Zoonotic Infections, Liverpool, UK. 306Centre for Clinical Infection and Diagnostics Research, Department of Infectious Diseases, School of Immunology and Microbial Sciences, King’s College London, London, UK. 307Department of Infectious Diseases, Guy’s and St Thomas’ NHS Foundation Trust, London, UK. 308Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, UK. 309Department of Pediatrics and Virology, St Mary’s Medical School Bldg, Imperial College London, London, UK. 310NHS Greater Glasgow & Clyde, Glasgow, UK. 311The Florey Institute for Host-Pathogen Interactions, Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, UK. 312Walton Centre NHS Foundation Trust, Liverpool, UK. 313MRC Centre for Molecular Bacteriology and Infection, Imperial College London, London, UK. 314Division of Structural Biology, The Wellcome Centre for Human Genetics, University of Oxford, Headington, Oxford, OX3 7BN, UK. 315Blood Borne Virus Unit, Virus Reference Department, National Infection Service, Public Health England, London, UK. 316Transfusion Microbiology, National Health Service Blood and Transplant, London, UK 317Department of Medicine, Imperial College London, London, UK. 318Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, UK. 319Tropical & Infectious Disease Unit, Royal Liverpool University Hospital, Liverpool, UK. 320Liverpool Clinical Trials Centre, University of Liverpool, Liverpool, UK. 321Centre for Health Informatics, Division of Informatics, Imaging and Data Science, School of Health Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester Academic Health Science Centre, Manchester, UK. 322Department of Molecular and Clinical Cancer Medicine, University of Liverpool, Liverpool, UK. 323Institute of Translational Medicine, University of Liverpool, Liverpool, Merseyside, UK.
The list of members and their affiliations can be found in the supplementary information.
HGI Consortium (Covid-19 Host Genetics Initiative)
Andrea Ganna218,219, Patrick Sulem220, David A. van Heel221, Mattia Cordioli218, Alessandra Renieri31,32, Gardar Sveinbjornsson220, Mari E. K. Niemi218 & Alex Pereira222
218Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland. 219Analytic & Translational Genetics Unit, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA. 220deCODE genetics/Amgen, Inc., Sturlugata 8, 101 Reykjavik, Iceland. 221Blizard Institute, Queen Mary University of London, 4 Newark Street, London, UK. 222Heart Institute, University of Sao Paulo, Brazil.
The list of members and their affiliations can be found in the supplementary information.
23andMe Contributors
Janie F. Shelton223, Anjali J. Shastri223, Chelsea Ye223, Catherine H. Weldon223, Teresa Filshtein-Sonmez223, Daniella Coker223, Antony Symons223 , Jorge Esparza-Gordillo224, The 23andMe COVID-19 Team223, Stella Aslibekyan223 & Adam Auton223
22323andMe Inc., 223 N Mathilda Ave, Sunnyvale, CA, 94086, USA. 224Human genetics - R&D, GSK Medicines Research Centre, Target Sciences-R&D, Stevenage, UK.
The list of members and their affiliations can be found in the supplementary information.
GEN-COVID Contributors
Francesca Mari31,32, Sergio Daga31, Margherita Baldassarri31, Elisa Benetti225, Simone Furini225, Chiara Fallerini31, Francesca Fava31,32, Floriana Valentino31, Gabriella Doddato31, Annarita Giliberti31, Rossella Tita32, Sara Amitrano32, Mirella Bruttini31,32, Susanna Croci31, Ilaria Meloni31, Anna Maria Pinto32, Elisa Frullanti31, Ilaria Meloni31, Caterina Lo Rizzo32, Francesca Montagnani226, Laura Di Sarno31, Andrea Tommasi31,32, Maria Palmieri31, Arianna Emiliozzi226, Massimiliano Fabbiani226, Barbara Rossetti226, Giacomo Zanelli226, Elena Bargagli227, Laura Bergantini227, Miriana D’Alessandro227, Paolo Cameli227, David Bennet227, Federico Anedda228, Simona Marcantonio228, Sabino Scolletta228, Federico Franchi228, Maria Antonietta Mazzei229, Susanna Guerrini229, Edoardo Conticini230, Luca Cantarini230, Bruno Frediani230, Danilo Tacconi231, Chiara Spertilli231, Marco Feri232, Alice Donati232, Raffaele Scala233, Luca Guidelli233, Genni Spargi234, Marta Corridi234, Cesira Nencioni235, Leonardo Croci235, Gian Piero Caldarelli236, Maurizio Spagnesi237, Paolo Piacentini237, Maria Bandini237, Elena Desanctis237, Silvia Cappelli237, Anna Canaccini238, Agnese Verzuri238, Valentina Anemoli238, Antonella D’Arminio Monforte239, Esther Merlini239, Mario U. Mondelli240,241, Stefania Mantovani240, Serena Ludovisi240,241, Massimo Girardis242, Sophie Venturelli242, Marco Sita242, Andrea Antinori243, Alessandra Vergori243, Stefano Rusconi244,245, Matteo Siano245, Arianna Gabrieli245, Agostino Riva244,245, Daniela Francisci246, Elisabetta Schiaroli246, Pier Giorgio Scotton247, Francesca Andretta247, Sandro Panese248, Renzo Scaggiante249, Francesca Gatti249, Saverio Giuseppe Parisi250, Francesco Castelli251, Maria Eugenia Quiros-Roldan251, Paola Magro251, Isabella Zanella252, Matteo Della Monica253, Carmelo Piscopo253, Mario Capasso254,255,256, Roberta Russo254,255, Immacolata Andolfo254,255, Achille Iolascon254,255, Giuseppe Fiorentino257, Massimo Carella258, Marco Castori258, Giuseppe Merla258, Filippo Aucella259, Pamela Raggi260, Carmen Marciano260, Rita Perna260, Matteo Bassetti261,262, Antonio Di Biagio262, Maurizio Sanguinetti263,264, Luca Masucci263,264, Serafina Valente265, Marco Mandalà266, Alessia Giorli266, Lorenzo Salerni266, Patrizia Zucchi267, Pierpaolo Parravicini267, Elisabetta Menatti268, Stefano Baratti269, Tullio Trotta270, Ferdinando Giannattasio270, Gabriella Coiro270, Fabio Lena271, Domenico A. Coviello272, Cristina Mussini273, Giancarlo Bosio274, Enrico Martinelli274, Sandro Mancarella275, Luisa Tavecchia275, Lia Crotti276,277, Nicola Picchiotti278,279, Marco Gori278,280, Chiara Gabbi281, Maurizio Sanarico282, Stefano Ceri283, Pietro Pinoli283, Francesco Raimondi284, Filippo Biscarini285,324 & Alessandra Stella285,323
225Department of Medical Biotechnologies, University of Siena, Siena, Italy. 226Dept of Specialized and Internal Medicine, Tropical and Infectious Diseases Unit, University Hospital of Siena, Siena, Italy. 227Unit of Respiratory Diseases and Lung Transplantation, Department of Internal and Specialist Medicine, University of Siena, Siena, Italy. 228Dept of Emergency and Urgency, Medicine, Surgery and Neurosciences, Unit of Intensive Care Medicine, Siena University Hospital, Siena, Italy. 229Department of Medical, Surgical and Neurosciences and Radiological Sciences, Unit of Diagnostic Imaging, University of Siena, Siena, Italy. 230Rheumatology Unit, Department of Medicine, Surgery and Neurosciences, University of Siena, Policlinico Le Scotte, Siena, Italy. 231Department of Specialized and Internal Medicine, Infectious Diseases Unit, San Donato Hospital Arezzo, Arezzo, Italy. 232Dept of Emergency, Anesthesia Unit, San Donato Hospital, Arezzo, Italy. 233Department of Specialized and Internal Medicine, Pneumology Unit and UTIP, San Donato Hospital, Arezzo, Italy. 234Department of Emergency, Anesthesia Unit, Misericordia Hospital, Grosseto, Italy. 235Department of Specialized and Internal Medicine, Infectious Diseases Unit, Misericordia Hospital, Grosseto, Italy. 236Clinical Chemical Analysis Laboratory, Misericordia Hospital, Grosseto, Italy. 237Department of Preventive Medicine, Azienda USL Toscana Sud Est, Arezzo, Italy. 238Territorial Scientific Technician Department, Azienda USL Toscana Sud Est, Arezzo, Italy. 239Department of Health Sciences, Clinic of Infectious Diseases, ASST Santi Paolo e Carlo, University of Milan, Milan, Italy. 240Division of Infectious Diseases and Immunology, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy. 241Department of Internal Medicine and Therapeutics, University of Pavia, Pavia, Italy. 242Department of Anesthesia and Intensive Care, University of Modena and Reggio Emilia, Modena, Italy. 243HIV/AIDS Department, National Institute for Infectious Diseases, IRCCS, Lazzaro Spallanzani, Rome, Italy. 244III Infectious Diseases Unit, ASST-FBF-Sacco, Milan, Italy. 245Department of Biomedical and Clinical Sciences Luigi Sacco, University of Milan, Milan, Italy. 246Infectious Diseases Clinic, Department of Medicine 2, Azienda Ospedaliera di Perugia and University of Perugia, Santa Maria Hospital, Perugia, Italy. 247Department of Infectious Diseases, Treviso Hospital, Local Health Unit 2 Marca Trevigiana, Treviso, Italy. 248Clinical Infectious Diseases, Mestre Hospital, Venice, Italy. 249Infectious Diseases Clinic, ULSS1, Belluno, Italy. 250Department of Molecular Medicine, University of Padova, Padua, Italy. 251Department of Infectious and Tropical Diseases, University of Brescia and ASST Spedali Civili Hospital, Brescia, Italy. 252Department of Molecular and Translational Medicine, University of Brescia, Italy; Clinical Chemistry Laboratory, Cytogenetics and Molecular Genetics Section, Diagnostic Department, ASST Spedali Civili di Brescia, Brescia, Italy. 253Medical Genetics and Laboratory of Medical Genetics Unit, A.O.R.N. “Antonio Cardarelli”, Naples, Italy. 254Department of Molecular Medicine and Medical Biotechnology, University of Naples Federico II, Naples, Italy. 255CEINGE Biotecnologie Avanzate, Naples, Italy. 256IRCCS SDN, Naples, Italy. 257Unit of Respiratory Physiopathology, AORN dei Colli Monaldi Hospital, Naples, Italy. 258Division of Medical Genetics, Fondazione IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo, Italy. 259Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo, Italy. 260Clinical Trial Office, Fondazione IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo, Italy. 261Department of Health Sciences, University of Genova, Genoa, Italy. 262Infectious Diseases Clinic, Policlinico San Martino Hospital, IRCCS for Cancer Research Genoa, Genova, Italy. 263Microbiology, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Catholic University of Medicine, Rome, Italy. 264Department of Laboratory Sciences and Infectious Diseases, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy. 265Department of Cardiovascular Diseases, University of Siena, Siena, Italy. 266Otolaryngology Unit, University of Siena, Siena, Italy. 267Department of Internal Medicine, ASST Valtellina e Alto Lario, Sondrio, Italy. 268Study Coordinator Oncologia Medica e Ufficio Flussi, Sondrio, Italy. 269Department of Infectious and Tropical Diseases, University of Padova, Padua, Italy. 270First Aid Department, Luigi Curto Hospital, Polla, Salerno, Italy. 271Local Health Unit-Pharmaceutical Department of Grosseto, Toscana Sud Est Local Health Unit, Grosseto, Italy. 272U.O.C. Laboratorio di Genetica Umana, IRCCS Istituto G. Gaslini, Genoa, Italy. 273Infectious Diseases Clinics, University of Modena and Reggio Emilia, Modena, Italy. 274Department of Respiratory Diseases, Azienda Ospedaliera di Cremona, Cremona, Italy. 275U.O.C. Medicina, ASST Nord Milano, Ospedale Bassini, Cinisello Balsamo, Italy. 276Istituto Auxologico Italiano, IRCCS, San Luca Hospital, Milan, Italy. 277Department of Medicine and Surgery, University of Milano-Bicocca, Milan, Italy. 278University of Siena, DIISM-SAILAB, Siena, Italy. 279Department of Mathematics, University of Pavia, Pavia, Italy. 280University Cote d’Azur, Inria, CNRS, I3S, Maasai, Nice, France. 281Independent Medical Scientist, Milan, Italy. 282Independent Data Scientist, Milan, Italy. 283Department of Electronics, Information and Bioengineering (DEIB), Politecnico di Milano, Milano, Italy. 284Scuola Normale Superiore, Pisa, Italy. 285CNR-Consiglio Nazionale delle Ricerche, Istituto di Biologia e Biotecnologia Agraria (IBBA), Milano, Italy. 324Present address: ERCEA (European Research Council Executive Agency), Bruxelles, Belgium.
The list of members and their affiliations can be found in the supplementary information.
BRACOVID Contributors
Alexandre C. Pereira222, Jose E. Krieger222, Emmanuelle Marques222 & Cinthia E. Jannes222
The list of members and their affiliations can be found in the supplementary information.
